SACR: The Swiss Ageing Citizen Reference
Main PI: Prof. Nicole Probst-Hensch (Swiss Tropical and Public Health Institute)
Project consortium: Prof. Murielle Bochud (Unisanté), Prof. Martin Preisig (CHUV), Prof. Peter Vollenweider (CHUV), Prof. Bogdan Draganski (CHUV), Prof. Oliver Bieri (University of Basel), Prof. Andreas Papassotiropoulos (University of Basel)
Project team: Dr. Ayoung Jeong (Swiss Tropical and Public Health Institute), Dr. Medea Imboden (Swiss Tropical and Public Health Institute), Dr. Jean-Pierre Ghobril (Unisanté), Sandrine Estoppey (Unisanté)
Main achievements
The SPHN SACR project aimed to
- Harmonize and apply data and biomaterial from existing Swiss citizen cohorts to build the well-embedded public health pillar of personalized health in Switzerland.
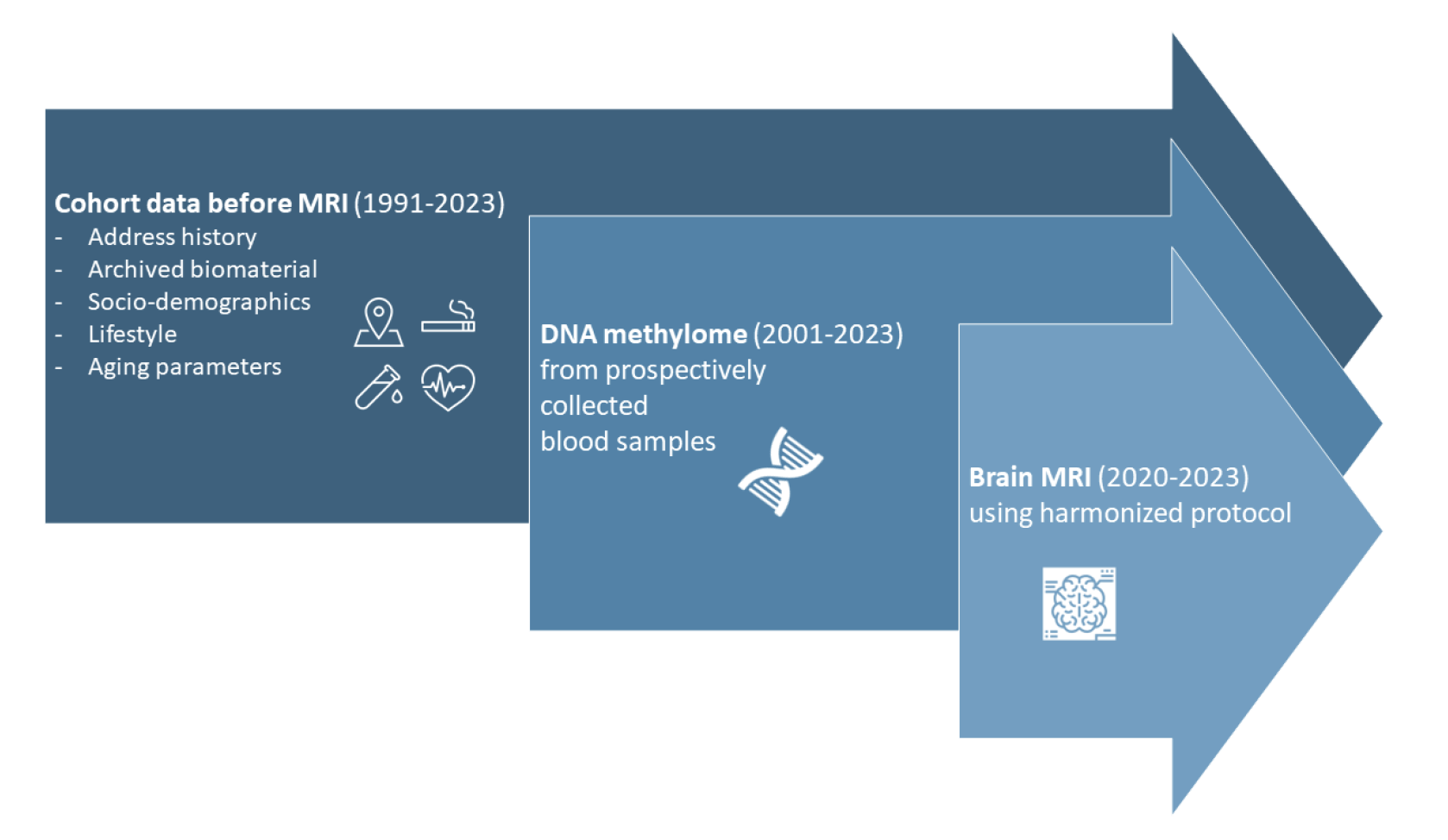
- Grant personalized health researchers responsible access to prospectively obtained information on health and health risks, to geocoded residential and work address history, and to archived biomaterial for testing the clinical and public health utility of candidate biomarkers and exposome features in predicting aging-related intermediate biomarkers (DNA methylation; MRI derived brain features), and subsequently aging-related (multi-) morbidities.
To achieve this goal, the project has been split into four Work packages: Infrastructure (WP1), Methylome analysis (WP2), MRI registry (WP3) and Scientific output (WP4).
SACR puts together three population-based cohorts (SAPALDIA, CoLaus/PsyCoLaus, and SKIPOGH) to create a novel data set of sample size >900, including brain MRI, DNA methylome of prospective biospecimen, and aging phenotypes. The SACR database will make a unique contribution to the aging research in itself or by contributing healthy reference data for case-control analysis.
Reusable datasets and infrastructures
Deep phenotyping in the context of previous cohort examinations
The SACR standard dataset was developed by matching variables from the population-based cohorts SAPALDIA, CoLaus/PsyCoLaus, and SKIPOGH.
Variables comprising personal characteristics, lifestyle and adiposity, and healthy aging related phenotypes have been assembled and harmonized across the three cohorts.
Available resources
How to get access to the SACR database?
Interested researchers can fill in the SACR research application form and submit it to nicole.probst@swisstph.ch.
The SACR consortium board will review the application and inform the applicant. Upon the acceptance of the application, project-specific DTA/MDTA will be prepared by SACR consortium. Costs for data access and analysis capacity may arise for the applicant.
Brain MRI features
902 participants of SACR have had their MRI taken at one of the SACR study centers. The image processing pipeline is similar to the one described by Trofimova et al. (Commun Biol. 2023 Apr 10;6(1):392) and transforms the raw voxel-based data into segmented data where standardized percentage intensity values are given per anatomical region and modality.
The anatomical region definitions are a custom selection of mesh terms, UMLS terms and anatomical regions defined in the neuromorphometrics challenge.
Available resources
DNA methylome
Blood samples mostly prospectively sampled from 891 out of the 902 participants with MRI have been identified. DNA methylome analysis is performed using the Illumina BeadChip Array (v2.0). Bisulfite treatment and methylome assessment are conducted on DNA samples in a randomized order, which minimizes batch effects. The randomization scheme has been described in detail in this document.
The DNA methylome data will be processed following the same principle as the previous DNA methylome analyses in SAPALDIA (Jeong et al 2022; Jeong et al 2019), including background and dye-bias correction, beta-mixture quantile normalization, and batch effect control.
Available resources
Planning SACR DNA Methylation Analysis.pdf
DNAmethylome_codebook (will follow in Q3 2024).
Data storage, access, organisation and documentation
A BioMedIT tenant for SACR-SPHN has been created for data storage, analysis, and manipulation. It can be accessed under https://portal.dcc.sib.swiss/.
Prerequisites are that the users are accredited members of SACR. External users can request access through the cohort access form. Upon approval, they will be given access to the tenant.
The SACR BioMedIT tenant comprises a remote desktop environment for file manipulation and remote work and data analysis instances. Remote Jupyter notebooks and remote R-Studio services have been implemented for data analysis purposes and can be launched onto variable power computing infrastructure depending on the needs of the end user. A GraphDB instance has also been created on the BiomedIT tenant and can be used to query the SACR data in graphical format.
All tools can be accessed under https://sphn-sacr-ondemand.scicore.unibas.ch/
All the cohort data has been uploaded onto the SACR BioMedIT tenant using the SETT secure encryption and transfer tool.
Available resources
Portal: https://portal.dcc.sib.swiss/.
Access type: SWITCHeduID + member of SACR for BiomedIT portal access
Remote services: https://sphn-sacr-ondemand.scicore.unibas.ch/
Access type: SWITCHeduID + member of SACR for BiomedIT portal access
Metadata: Maelstrom catalogue
Access type: Public
Further links and references
Note that the SACR cohorts, which have been implemented with different objectives, have much richer information over longer time period, while the SACR codebook contain a more limited, but fully harmonized number of variables.
The whole spectrum of information can be explored via meta-data databases:
References on Pubmed
- Jeong A, Bochud M, Cattin P, et al. SPHN - The Swiss Aging Citizen Reference (SACR). Studies in Health Technology and Informatics. 2020 Jun;270:1168-1169. DOI: 10.3233/shti200347. PMID: 32570565.
Disclaimer: The contents on this website are intended as a general source of information and have been provided by the project PIs. The SPHN Management Office is not responsible for its accuracy, validity, or completeness.

