QA4IQI - Quality assessment for interoperable quantitative CT-Imaging
Project consortium: Bram Stieltjes (USB), Adrien Depeursinge (HES-SO), Henning Müller (HES-SO), Ender Konukoglu (ETH), Hatem Alkadhi (USZ), Clarisse Dromain (CHUV), Hendrik von Tengg-Kobligk (Inselspital Bern)
Additional contributors: Kyriakos Flouris (ETH), Oscar Jimenez-del-Toro, Roger Schaer (HES-SO), Pierre-Alexandre Poletti (HUG), Rolf Hügli (KSBL), Christoph Aberle, Michael Bach, Markus M Obmann (USB).
Further supporting institutions: 3R - Réseau Radiologique Roman, Schmerzklinik Basel, Zentrum für Bilddiagnostik Basel
Main achievements
Phantom - Development
An anthropomorphic CT phantom has been developed. It highly realistically simulates the attenuation properties of human tissue. Figure 1 on the left shows the data set of an abdominal CT examination, and on the right, the corresponding CT measurements of the phantom. The red rectangle indicates the printable area (DIN A4). In the range of typical soft tissue HU values, there are hardly any visible differences between the phantom and a real patient examination. This is made possible by printing an iodine-containing ink on paper with an inkjet printer. The phantom can thus be built up layer by layer or in other words paper sheet by paper sheet. A previously acquired CT data set or artificially generated images can serve as a template.
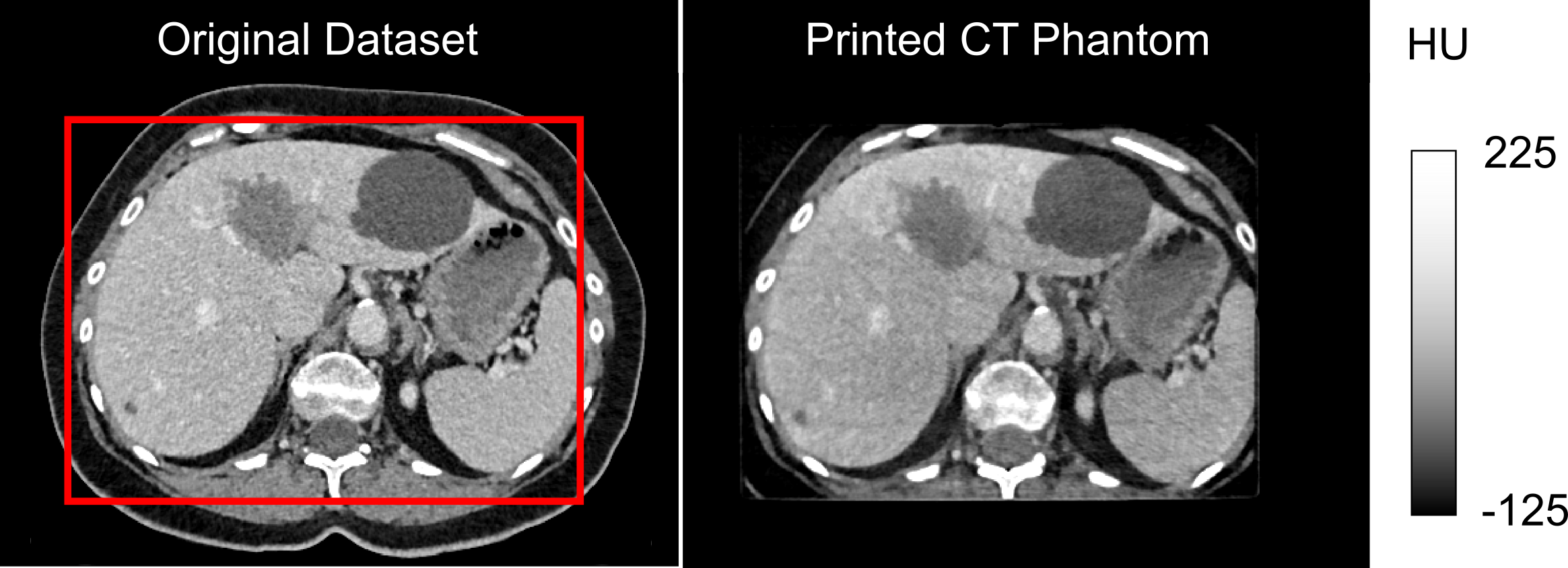
Figure 1: Real patient examination (left) in comparison to phantom measurement (right).
Multicenter data set
The phantom allows realistic measurements of the same "patient" at different CT scanners and locations. Measurements have been made across Switzerland on 13 different CT scanners and all common manufacturers. The CT dataset is made available to the community.
Pipeline for automatic evaluation of image data sets
The acquired data sets were processed according to the flowchart shown in figure 2. After measuring the phantom in the CT scanner, the data was first pseudonymized and then uploaded to the Kheops platform. From here, further processing could take place. In our project, the ROIs once defined in the phantom were now registered to each newly acquired dataset. Image features (radiomics) were then extracted. The pipeline can be used in general for image feature extraction as long as a dataset and the corresponding ROIs are available. The pipeline is provided as open source.
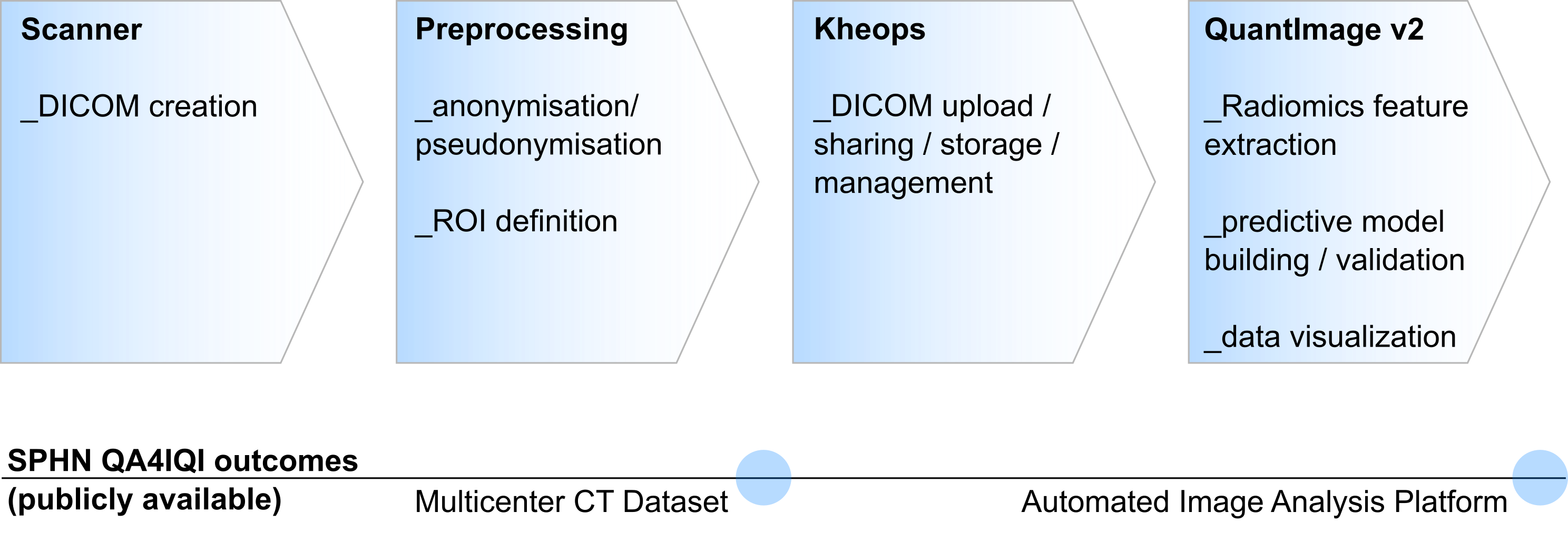
Figure 2: Image analysis pipeline and related SPHN QA4IQI outcomes.
Reusable infrastructures and datasets
Standardized CT Multicenter Dataset
The dataset contains highly anthropomorphic CT phantom images from the thorax, the abdomen, and test patterns (see figure 3). The datasets were acquired in a standardized manner at 8 institutions (5 University Hospitals and 3 other imaging centers) and in total at 13 CT machines. All common vendors are represented (Siemens, Philips, Canon/Toshiba, GE). The dataset can be used, for example, to compare biomarkers or quantify their expected variations. This helps in the design of clinical trials and the development of new biomarkers that potentially are used in the future at several stages of patient care: for diagnosis, staging, and prognosis, as well as for predicting and detecting treatment response.
Available resources
The data are accessible through the Kheops platform (https://kheops.ehealth.hevs.ch). Publication on The Cancer Imaging Archive (TCIA) is ongoing. The awareness of the dataset is also raised by further scientific publications that are work in progress and refer to the dataset (e.g. nature scientific data). A further source of information is the project webpage (https://qa4iqi.github.io/).
The data are available in the DICOM format – the standard for the communication and management of medical imaging information.
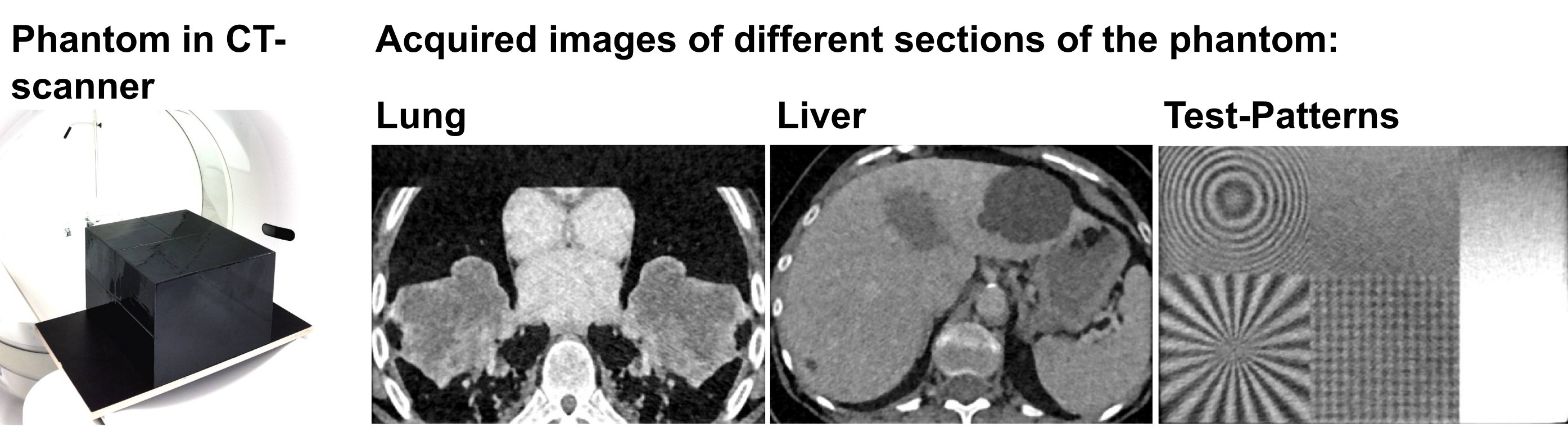
Figure 3: CT Phantom and its different sections after image acquisition.
Automated image analysis platform
An analysis pipeline was created to evaluate the datasets. It enables the standardized evaluation of all datasets. All necessary tools are openly accessible in a GitHub repository and can freely be used by the community. Thus, the analysis can easily be extended with any other imaging biomarker or used for other datasets.
The efforts of QA4IQI contributed to the development of the QuantImage v2 platform. The platform allows clinical researchers with no programming background to rapidly investigate the relevance of radiomics models in a novel application context. QuantImage v2 is publicly available along with the instance of Kheops that was used to share the data in this project. In particular, shared components for feature extraction between the QA4IQI pipeline and the QuantImage v2 platform were developed.
Available resources
Github: https://github.com/QA4IQI
Kheops Platform: https://kheops.ehealth.hevs.ch
QuantImage v2 Platform: https://quantimage2.ehealth.hevs.ch/
Follow-up projects - continuation - next steps
The data sets generated have already led to initial results and publications during the project period, but by no means have all the findings that can be derived from the existing data been processed. While so far mainly the dependency of the biomarkers on the acquisition parameters has been investigated, the focus is now on the dependency on different device manufacturers and device versions, which is especially important for multi-center studies. In addition, the creation of further data sets with the phantom is planned. The infrastructure produced in this project will be used and further developed. The resulting software updates will be made available to the community via the GitHub page.
Links and references relevant to the project
- Project website: https://qa4iqi.github.io/
- Code on Github: https://github.com/QA4IQI/qa4iqi-extraction
- Kheops Platform: https://kheops.ehealth.hevs.ch
- QuantImage v2 Platform: https://quantimage2.ehealth.hevs.ch/
Further information
Phantom Development: PhantomX, https://phantomx.de
Watch the SPHN webinar
Presenter: Dr Bram Stieltjes, Vice Chair of Research, Radiology, Head of Research and Analytics, ICT, University Hospital Basel.
References on Pubmed
- Jimenez-Del-Toro O, Aberle C, Bach M, Schaer R, Obmann MM, Flouris K, Konukoglu E, Stieltjes B, Müller H, Depeursinge A. The Discriminative Power and Stability of Radiomics Features With Computed Tomography Variations: Task-Based Analysis in an Anthropomorphic 3D-Printed CT Phantom. Invest Radiol. 2021 Dec 1;56(12):820-825. doi: 10.1097/RLI.0000000000000795. PMID: 34038065.
- Flouris K, Jimenez-Del-Toro O, Aberle C, Bach M, Schaer R, Obmann MM, Stieltjes B, Müller H, Depeursinge A, Konukoglu E. Assessing radiomics feature stability with simulated CT acquisitions. Sci Rep. 2022 Mar 18;12(1):4732. doi: 10.1038/s41598-022-08301-1. PMID: 35304508; PMCID: PMC8933485.
Disclaimer: The contents on this website are intended as a general source of information and have been provided by the project PIs. The SPHN Management Office is not responsible for its accuracy, validity, or completeness.

