SVIP-O: the Swiss Variant Interpretation Platform for Oncology
Project consortium: Daniel Stekhoven (ETHZ), Patrick Ruch (HES-SO), Valérie Barbié (SIB), Bernd Rinn (ETHZ), Ioannis Xenarios (SIB), Michael Baudis (UZH), Vincent Zoete (UniL), Alan Bridge (SIB).
In addition to the SPHN funding, SVIP-O also received funding from BioMedIT.
Main achievements
The Swiss Variant Interpretation Platform for Oncology (SVIP-O) has developed a framework to enable a one-stop shop for variant interpretation in cancer diagnostics (Figure 1). It has been specifically developed with Swiss pathologists, oncologists and haemato-oncologists to address the current needs in their fields. The three main aspects are:
- single point for comprehensive annotations across different resources;
- harmonized annotations of variants for all hospitals; and
- clinically validated interpretation of clinical significance.
SVIP deals with these aspects using a web-based frontend for retrieval, curation and expert review. In addition, a secure transaction space has been developed to enable query-based reporting, matchmaking of cases, and notifications of changed variant annotations. This secure instance is based at the SIB SENSA platform and has been objected to penetration tests.
In addition, a process to curate variants for cancer has been conceived and served as a basis to develop the curation interface, which is fully implemented and is delivering SVIP-specific annotations. The review interface is implemented, but has not yet been tested with the clinical expert panel.
The curation process is supported by tools to mine the literature and provide score-based prioritization of publications with respect to gene, variant, and disease type. Furthermore, the text mining uses term expansion to cover different ways to naming variants and genes to maximize recall.
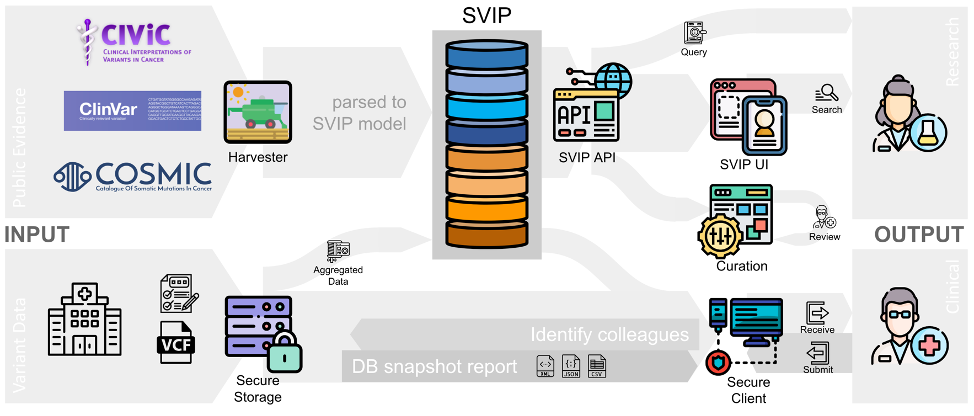
Figure 1: The SVIP-O framework as one-stop shop for variant interpretation in cancer diagnostics.
Reusable infrastructure and datasets
Public resource of variant annotations (www.svip.ch)
This infrastructure component of SVIP has two flavors: the public resource which is available at www.svip.ch containing the variant annotation and the secure infrastructure at SIB SENSA (https://sensa.sib.swiss/) enabling the secure handling of data from the partnering hospitals.
Harmonized variant annotations are published on www.svip.ch and can be queried through the stable API at https://api.svip.ch. The SVIP API is documented on Swagger. The secure infrastructure component has one access point for data providers and curators, but the two roles have different access rights (Figure 2).
Available ressources
- SVIP - Swiss service - https://svip.ch
- Stable API to SVIP - https://api.svip.ch
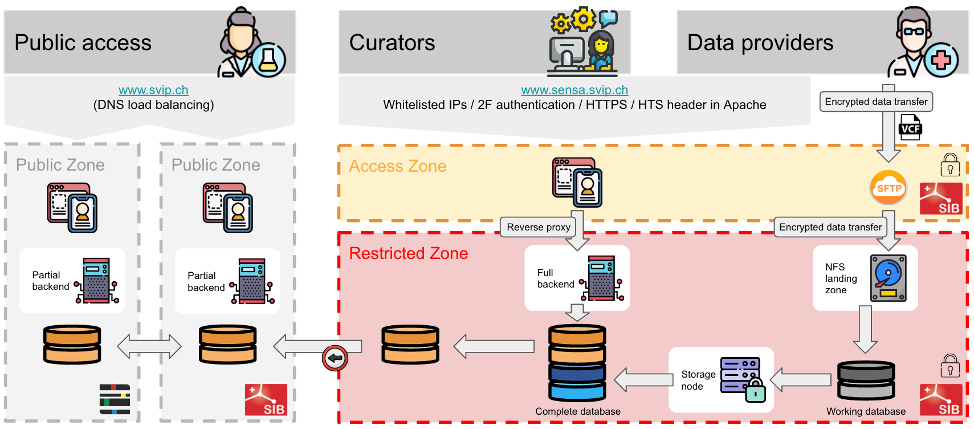
Figure 2: SVIP-O as public resource of variant annotations and secure transaction space for data providers.
Source code of SVIPdb on Github
The source code of SVIP is publicly available on Github. It contains the complete frontend and backend, which we call SVIPdb. This includes the interfaces for the public, curators and reviewers. SVIPdb enables to set up your own SVIP instance and make use of the curation and review functionalities on your own variant data.
To request new features or report bugs, issue templates are directly available through GIthub and enable the community to feedback on their experience with the SVIPdb code.
Available ressources
- SVIPdb - source code (GPL 3.0) - https://github.com/SVIPdb
- Feature requests/bug reports: https://github.com/SVIPdb/coordinator/issues
Data dictionaries
SVIP is utilizing established standards and ontologies, whenever possible. A comprehensive list can be found on www.svip.ch/help
Follow up projects – continuation – next steps
SVIP aims to become a reference for interpretation of variants in cancer in Swiss diagnostic routine, while at the same time being ready for research and development in the domain of Personalized Health. For the future, we plan to focus on three main aspects:
- Establishing Clinical Utility
We want to demonstrate how SVIP can support and improve clinical diagnostics at Swiss hospitals. This will start with well controlled tests in molecular labs, willing to try SVIP alongside their standard tools. Based on their feedback, we will adapt and extend the service to meet the needs of our partners. - Expand Global Collaboration
SVIP has built a good relationship with existing stakeholders in the domain of variant interpretation. Most notably, SVIP has been closely working with GA4GH VICC to adopt variant representation standards and also aims to use new versions of Beacon in the future. Community-driven coding and use of established standards will further promote excellence. - Curation & New Therapeutic Areas
Built on needs of the medical community, the SVIP project may expand its tools/expertise on curation & validation to other areas. The secure infrastructure setup is ready for medical application and can be specifically developed and extended to meet the needs of physicians & patients as required.
Links and reference to the project
- SVIP Intro Slide Deck (http://bit.ly/SVIP_intro)
- SVIP Documentation (https://www.svip.ch/help)
- SVIP on Expasy (https://www.expasy.org/resources/svip)
Further information
- Variomes (https://candy.hesge.ch/Variomes/)
- MetaKB (https://search.cancervariants.org/)
- GA4GH VICC (https://cancervariants.org/)
- Swiss-Prot (https://www.sib.swiss/swiss-prot)
- SwissPO (https://swiss-po.ch/)
Watch the SPHN Webinar
SVIP-O aims to set up a single place for genetic mutations in cancer identified from Swiss hospitals' patients. This curated database will enable clinicians to base their diagnoses on the same harmonized high-quality information, exchange among themselves on potential treatments, and get notified on changes in annotation.
References on Pubmed
- Stekhoven Daniel J., Ruch Patrick, Barbié Valérie. Swiss Variant Interpretation Platform for Oncology (SVIP-O). Swiss Med Informatics. 2018;34:00411
- Déborah Caucheteur, Emilie Pasche, Julien Gobeill, Anaïs Mottaz, Luc Mottin and Patrick Ruch. Designing retrieval models to contrast precision-driven ad hoc search vs. recall-driven treatment extraction in Precision Medicine. TREC 2019.
- Déborah Caucheteur, Julien Gobeill, Anaïs Mottaz, Emilie Pasche , Pierre-André Michel, Luc Mottin, Daniel J. Stekhoven, Valérie Barbié and Patrick Ruch. Text-mining Services of the Swiss Variant Interpretation Platform for Oncology. MIE 2020.
- Pasche et al 2021. Variomes: a high recall search engine to support the curation of genomic variants. May 2021. DOI: 10.1101/2021.05.29.446224
Disclaimer: The contents on this website are intended as a general source of information and have been provided by the project PIs. The SPHN Management Office is not responsible for its accuracy, validity, or completeness.

