The SPHN Semantic Interoperability Framework
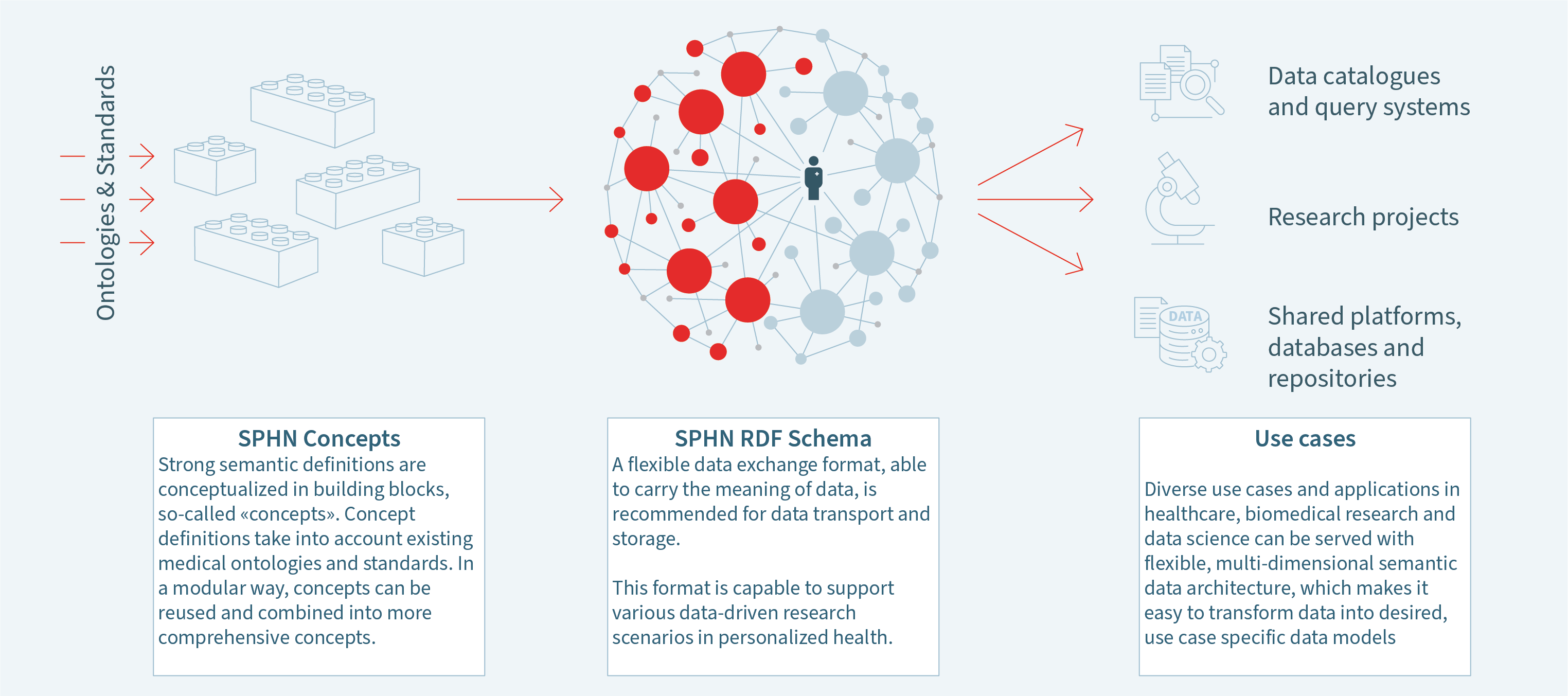
Health-related data, ranging from hospital routine data to biomedical research data, are quite diverse, stored in different databases and formats, and often coded in local standards. This fragmentation and diversity make it very time-consuming to combine data from different sources for doing research on a particular topic. It is generally difficult to understand data and the intended meaning due to the lack of common standards, metadata, or a common data dictionary. Following the FAIR (findable, accessible, interoperable and reusable) principles, SPHN builds an infrastructure to overcome these hurdles and enables collaborative research by making the meaning of health-related data understandable to humans and machines. This allows for an easy combination of the data from different sources, thus simplifying the use and exploration of data across Switzerland. Experience shows that the diverse needs and requirements of the different use cases make it impossible to agree on one single data model. Not only the required data elements differ between projects, but also the level of information granularity required for a single datapoint can vary significantly. Therefore, SPHN developed a framework based on a strong semantic layer of information (pillar 1), and graph technologies for the exchange layer, which can be extended by the individual projects to fit their purposes (pillar 2). Thus, a universal exchange language for healthcare is established, using the "words" from various international standard vocabularies (such as SNOMED CT or LOINC), a simple "grammar" (subject-predicate-object; expressed in RDF), and additional SPHN guidelines and rules to establish good practices for FAIR data.
How it is implemented
Semantic representation (Pillar 1)
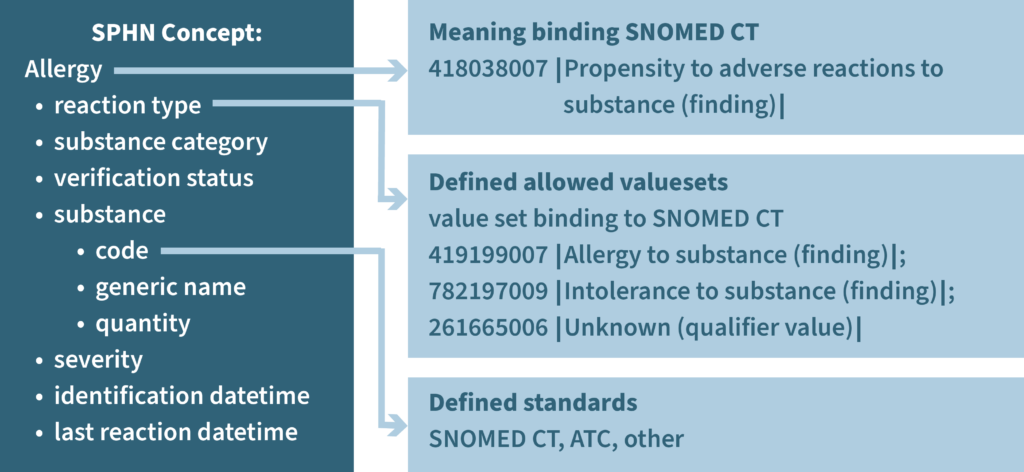
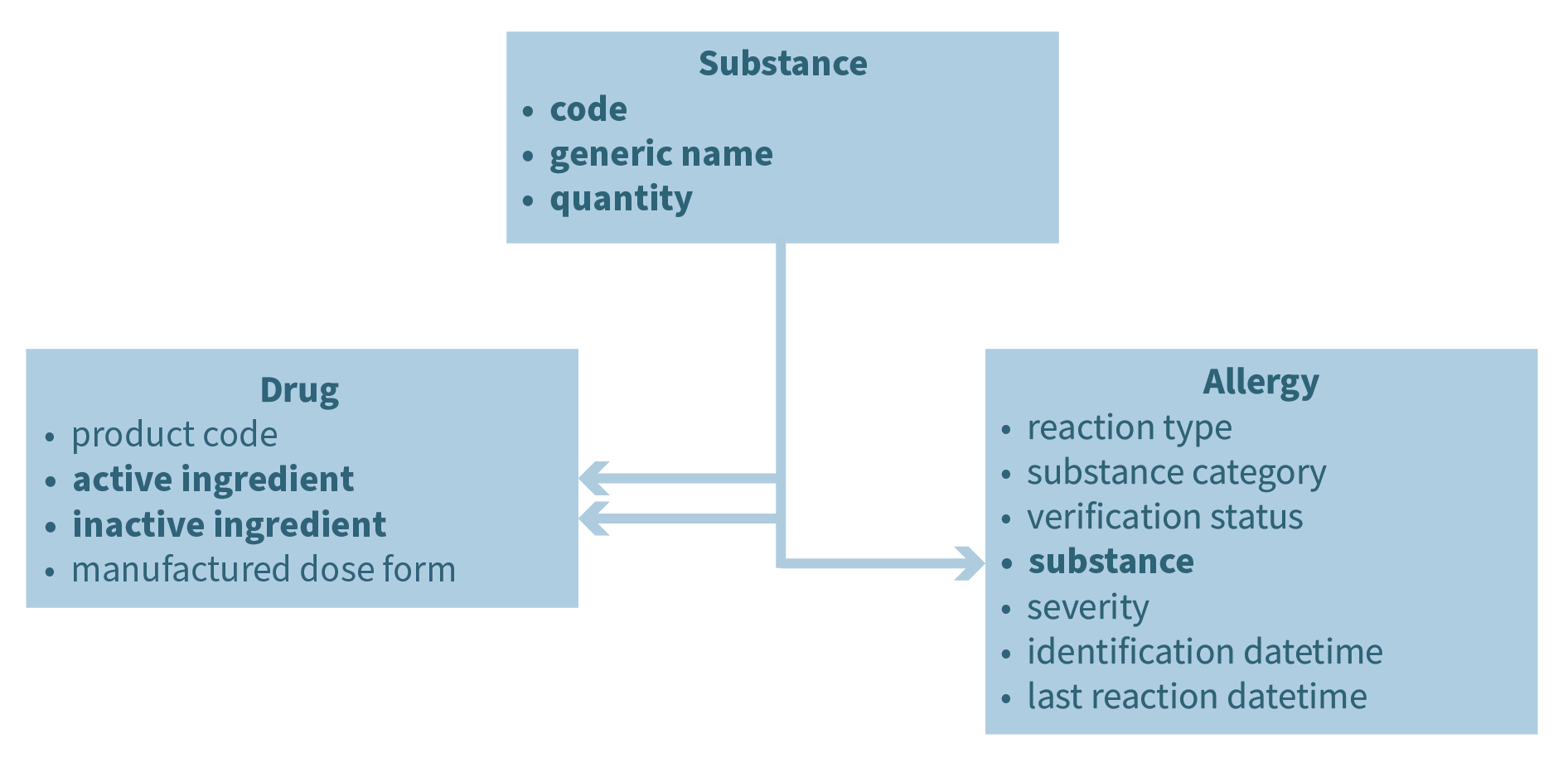
SPHN concepts are generalizable building blocks, which can be used in different contexts. Each concept contains all information necessary to understand it, and concepts can be combined to composed concepts, which again can be combined to more complex compositions. It is important to find the right level between abstraction and granularity to optimize the power of expression. The approach can be illustrated with the example of “substance”. A substance can be an active or inactive ingredient of a drug or it can be the substance someone is allergic to. Therefore, we can abstract “substance” as a concept on its own. The concept of substance is composed of three concepts: “code”, “generic name” and “quantity”. These concepts describe a substance no matter if it is the active ingredient of a drug (that is defined as the type “substance”) or the substance to which someone is allergic to.
To make the SPHN concepts comparable nationally and internationally, we express their meaning using existing semantic standards (controlled vocabularies), by creating a meaning binding wherever possible to SNOMED CT and/or LOINC. The data element of a concept can be expressed using one or several recommended standards (e.g. LOINC, SNOMED-CT, ICD-10, ICD-O-3, CHOP, ATC). For example, the instance of substance code under the concept “allergy” can be an ATC code, a SNOMED CT code, or a code form another semantic standard. If needed value sets are defined and if possible, a value set binding to SNOMED CT is added. Descriptions for concepts and value sets as well as standards are, whenever possible, aligned with national and international sources.
All SPHN concepts are represented in the SPHN Dataset, for the latest version click here.
Data transport and storage (Pillar 2)
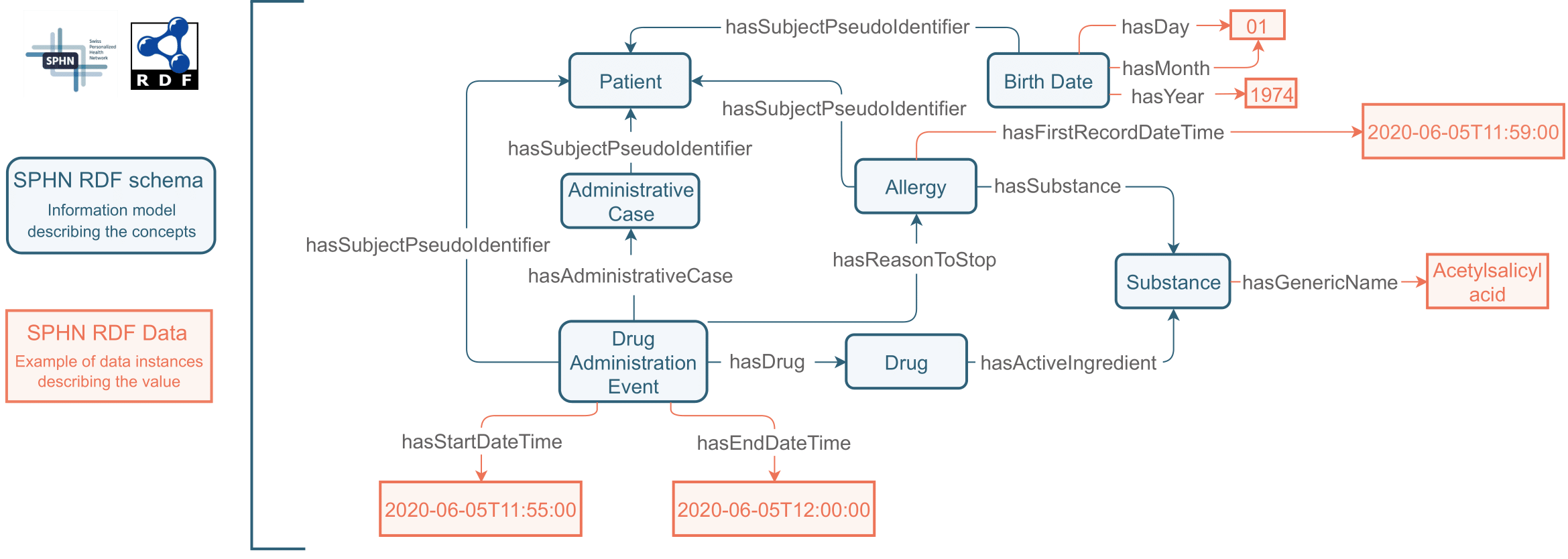
SPHN concepts (blue) and their instances (orange) can easily be mapped from/to other data representations or merged with other RDF data sets without losing their semantics.
The relations between different concepts and the data are expressed in the form of triples composed of a “subject”, a “predicate”, and an “object”. The example shows a patient born on the 1st of January 1974 which has an Administrative Case registered with a Drug Administration Event. During this administration, a Drug was given to the patient on the 05.06.2020 at 11:55. The Substance administered was 'Acetylsalicyl acid'. The Drug Administration Event has stopped five minutes later, at 12:00 for the reason that an Allergy was observed at 11:59 to that Substance. RDF is quite flexible and enable us to make use of different existing semantic standards, and value sets as defined in the SPHN Dataset.
In RDF subjects, predicates and (sometimes) objects have a Unique Resource Identifier (URI) that enables the unique identification of these elements. In our example, the concept Allergy has the URI https://biomedit.ch/rdf/sphn-ontology/sphn/Allergy , which uniquely and unambiguously identifies it in the context of SPHN. The SNOMED CT meaning binding to the equivalent meaning of ‘Allergy’ is introduced by linking the URI of the SPHN Allergy class to the corresponding URI in SNOMED CT (http://snomed.info/id/418038007) in the SPHN RDF schema.
Note that both URIs are resolvable on the web and can be browsed in their respective "ontologies”.
To facilitate the integration and therefore the use of external standards in the SPHN RDF schema and its related data, the DCC provides RDF files of different versions of the following coding systems: ATC, CHOP, LOINC, SNOMED CT, ICD-10-GM, UCUM.
Use cases (Pillar 3)
Based on these needs, researchers can:
- use the RDF files directly as input into their analysis software, e.g. rdflib in Python
- extract data into a flat file, e.g. Excel or CSV
- load the data into other existing data models with adequate converters, e.g. i2b2, OMOP or a data management software
How does this strategy help to address the following FAIR criteria:
Findable
- F1. (Meta)data are assigned a globally unique and persistent identifier
--> SPHN classes and properties are defined with URIs in the following namespace https://biomedit.ch/rdf/sphn-ontology/sphn#
--> data/resources use the following namespace https://biomedit.ch/rdf/sphn-resource/ and must be identified with a unique identifier for distinct data points
- F2. Data are described with rich metadata
--> each SPHN concept contains a set of information to describe the data element
--> administrative metadata (e.g., date of extraction, data provider who generated the files) is included directly in the RDF data files - F3. Metadata clearly and explicitly includes the identifier of the data they describe
--> data and metadata are linked via RDF properties - F4. (Meta)data are registered or indexed in a searchable resource
--> the SPHN RDF schema is openly available and searchable on the web (biomedit.ch)
Accessible
- A1. (Meta)data are retrievable by their identifier using a standardized communications protocol
--> the SPHN RDF schema URIs are resolvable on the web (accessible via https://)
--> SPARQL is the official RDF Query Language which is a standard protocol of the World Wide Web Consortium (W3C). It facilitates the exploration of data in RDF
Interoperable
- I1. (Meta)data use a formal, accessible, shared, and broadly applicable language for knowledge representation
--> RDF is the standard language used for knowledge representation - I2. (Meta)data use vocabularies that follow FAIR principles
--> SNOMED CT and LOINC along with other standards are used as controlled vocabulary in the SPHN RDF schema and in the data itself - I3. (Meta)data include qualified references to other (meta)data
--> when possible, the SPHN URIs of concepts and instances are linked to the URIs of the external resources like SNOMED CT, LOINC, ATC, ICD-10.
Reusable
- R1. (Meta)data are richly described with a plurality of accurate and relevant attributes
--> the SPHN RDF schema is published under the CC BY 4.0 licence
Watch the following videos to learn more about the SPHN Semantic Strategy
For more details see
- Semantic strategy developed by the Clinical Data Interoperability Semantic Expert Working Group
- Semantic Strategy Fact-Sheet
- SPHN Dataset with the released concepts
- Fact sheet on RDF
- SPHN RDF schema
- SPHN DCC Training
-
Documentation and User guide (ReadTheDocs)
- Progress Report on the RDF Roadmap
SPHN Interoperability Framework on PubMed
-
- Unni, D.; Touré, V.; Krauss, P.; Crameri, K.; Österle, S. SPHN Strategy to Unravel the Semantic Drift Between Versions of Standard Terminologies (doi:10.20944/preprints202312.0508.v1)
-
- van der Horst, E.; Unni, D.; Kopmels, F.; Armida, J.; Touré, V.; Franke, W.; Crameri, K.; Cirillo, E.; Österle, S. Bridging Clinical and Genomic Knowledge: An Extension of the SPHN RDF Schema for Seamless Integration and FAIRification of Omics Data (doi:2023120373)