SwissGenVar – A platform for clinical grade interpretation of genetic variants to foster personalized health care in Switzerland
Project consortium: Anita Rauch (University of Zurich), Sven Cichon (University Hospital and University of Basel), André Schaller (Bern University Hospital and Bern University), Marc Abramowicz (University Hospital and University of Geneva), Andrea Superti-Furga (University Hospital and University of Lausanne), Wolfgang Berger (University of Zürich), Valérie Barbié (SIB)
Additional persons involved: Dennis Kraemer (University of Zurich), Fabienne Maurer (University Hospital and University of Lausanne), Isabel Filges (University Hospital and University of Basel), Thierry Nouspikel (University Hospital and University of Geneva), Samuel Koller (University of Zurich), Stephanie Meier (University Hospital and University of Basel), Dillenn Terumalai (SIB), Javier Sanz (Bern University Hospital and Bern University), Livia Famiglietti (SIB).
Main achievements
The main achievement of this infrastructure development project is the establishment of the data sharing platform SwissGenVar, providing an environment for expert knowledge-sharing of genetic variant data to harmonize and up-scale their significance interpretation at clinical-grade and detailing available evidence following the international standards.
The SwissGenVar platform is running on the BioMedIT infrastructure, and allows for:
- data collection via a secure encrypted data transfer
- variants harmonized annotation
- secure data access and query by the project partners
and -for the first time ever-: connecting all Academic Centers of Human Genetics and Medical Genetics in Switzerland.
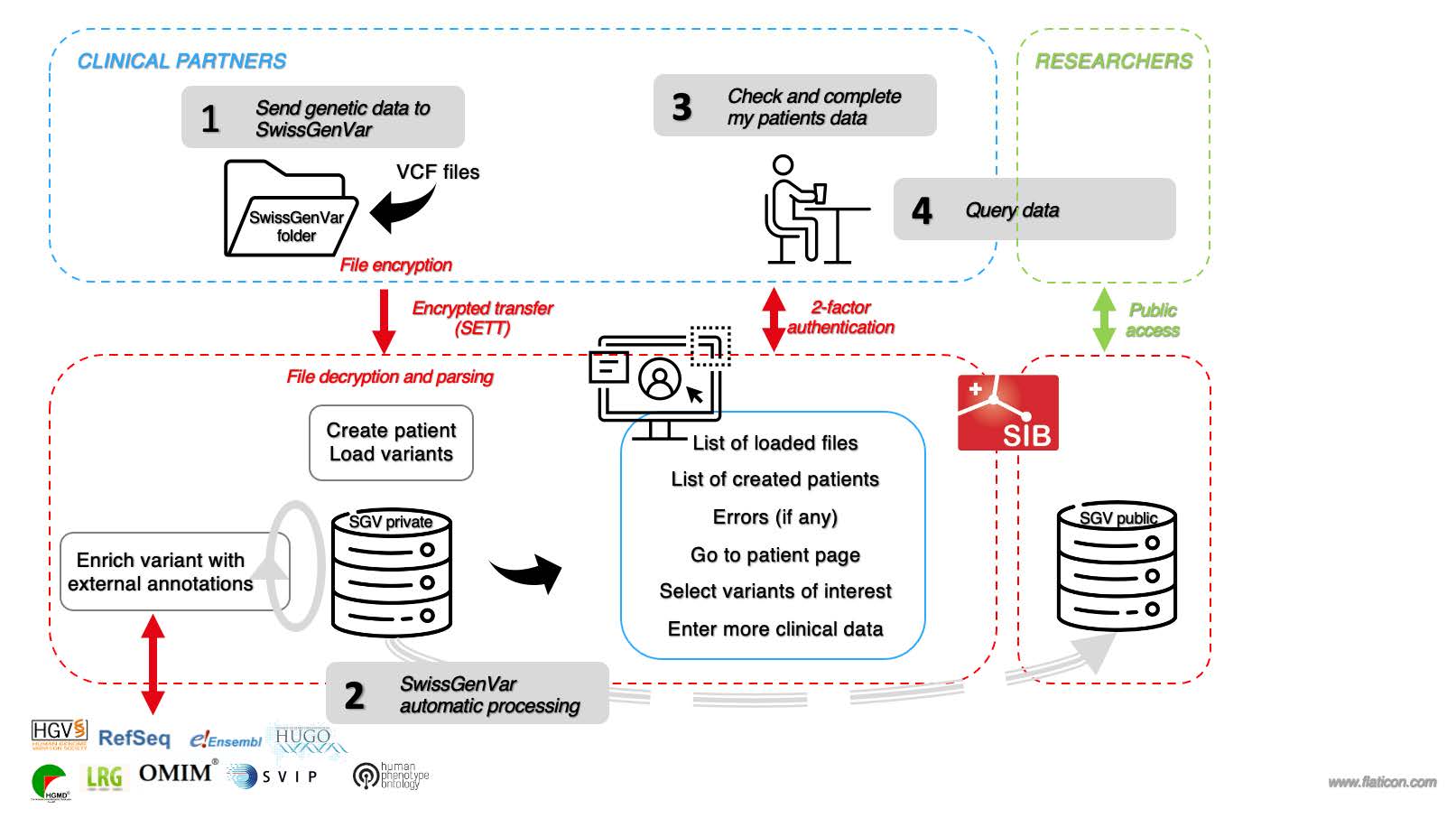
Figure 1: SwissGenVar application workflow
The Clinical Partners provide the genetic data to SwissGenVar in VCF format. The data are encrypted and securely transferred (step 1) using the SPHN data transfer tool.
Upon reception, the data are decrypted and loaded into the platform. The variants are automatically annotated using selected data sources (step 2).
At this stage, the Clinical Partners can connect to their protected account using 2-factor authentication to check the transfer of their data files and start adding clinical information on their patients (step 3). They can also query the full database using pre-defined filters (step 4).
In a second step, SwissGenVar will also encompass a publicly available platform where researchers will be able to access aggregated information.
The implementation of SwissGenVar is governed by the:
- SwissGenVar Consortium Agreement (CA)
- Data Transfer and Use Agreement (DTUA), which includes the SPHN Biomed IT Infrastructure Data Transfer and Processing Agreement (DTPA)
- Patients’ Consent Form (PCF)
Compliance with the Human Research Act (HRA) and General Data Protection Regulations (GDPR) as well as the guidelines of the SPHN ELSI Group are fully ensured.
Reusable infrastructures and datasets
Standardized Dataset Specifications
Consented high-quality basic clinical information accompanies the variant-related data to corroborate their diagnostic utility. The team defined a minimal and an extended genetic and clinical data set pertinent for variant data sharing/storing and clinical interpretation, in the current platform version only the core dataset is implemented
Whenever applicable, the existing international and/or SPHN standards are used in SwissGenVar. When there was no satisfying data standard available, the SwissGenVar consortium has adapted or created those relevant to the project. The data catalog was provided to the DCC for inclusion in the SPHN core data set.
Relational Database
An original database, able to handle patient- and variant-related data for depository and sharing of genetic variants and their clinical interpretation at diagnostic-grade, was developed. The data model is freely available.
Available resources
- The data model is available from the project public documentation page.
VCF Files Loader
A VCF file loader has been developed and optimized for a high-performance computing cluster (SLURM).
Available resources
- The tool is currently available upon request.
VCF Files Loader
A VCF file loader has been developed and optimized for a high-performance computing cluster (SLURM).
Available resources
- The tool is currently available upon request.
SwissGenVar Legal Documents
The SwissGenVar Consortium Agreement (CA) and the Data Transfer and Use Agreement (DTUA), which includes the SPHN Biomed IT Infrastructure Data Transfer and Processing Agreement (DTPA), have been established (with the SIB Lausanne BiomedIT node as host).
The Consortium Agreement contains:
- Database Access
- Governance Policies
- Data Transfer And Use Agreement (DTUA)
- Template Description For Future Projects Involving SwissGenVar
- Authorship Guidelines
The compliance with the HRA (Human Research Act) and GDPR (General Data Protection Regulations) as well as the guidelines of the SPHN ELSI group is fully ensured.
Available resources
- All schedules and templates are available upon request.
Patients’ Consent Form (PCF)
Based upon an existing common patient consent form of the Swiss Society of Medical Genetics and the recommended templates for general consents,
the consortium agreed upon a renewed Patients’ Consent Form (PCF) in line with the objectives of SwissGenVar in sharing and storing of at least a minimal set of genetic and related clinical data.
Available resources
- Document available upon request.
Data dictionnaries
Whenever applicable, SwissGenVar uses the existing international and/or SPHN standards ("Pre-existing catalogs and data sources"). When there was no satisfying data standard available, the SwissGenVar consortium has adapted or created those relevant to the project ("Internal catalogs").
Follow-up projects - continuation - next steps
The SwissGenVar Consortium has committed itself to sustain the infrastructure with institutional funds beyond the funding period of SPHN through its added value for routine diagnostics and research.
To foster personalized health research, we have submitted the SwissClinGen National Data Stream Proposal which is based on the SwissGenVar Platform.
The goal of SwissClinGenNDS is to create a framework that allows to connect clinical data from the University Hospital data warehouses with consented genetic data from routine diagnostics or research genetic testing hosted within the SwissGenVar Platform. This will allow to investigate in an unbiased and longitudinal manner the relation between genotypes and phenotypes and to study natural history of genetic disease predispositions and Mendelian or near-Mendelian disorders.
Links and references relevant to the project
- SwissGenVar Documentation: https://pages.sib.swiss/project/swissgenvar-doc/
- Public Website: https://pages.sib.swiss/project/swissgenvar/
- Project Gitlab: https://gitlab.sib.swiss/clinbio/swissgenvar/sgv-knowledge/-/tree/master
Further information
The SwissGenVar project has strongly contributed to harmonize practices between the participating institutions. The vocabularies and ontologies have been standardized after many rounds of discussion and were provided to the SPHN DCC to serve as a basis for other medical genetics projects.
We have implemented SPHN tools and vocabularies whenever possible, and are discussing with the DCC to identify the best tools and means to implement RDF, in collaboration with the SVIP-O project.
References on Pubmed
- Kraemer D, Terumalai D, Famiglietti ML, Filges I, Joset P, Koller S, Maurer F, Meier S, Nouspikel T, Sanz J, Zweier C, Abramowicz M, Berger W, Cichon S, Schaller A, Superti-Furga A, Barbié V, Rauch A. SwissGenVar: A platform for clinical grade interpretation of genetic variants to foster personalized health care in Switzerland.
Disclaimer: The contents on this website are intended as a general source of information and have been provided by the project PIs. The SPHN Management Office is not responsible for its accuracy, validity, or completeness.

