Population-wide screens of the immune repertoire: a reverse personalized-medicine approach
Project consortium: Prof. Adriano Aguzzi (University of Zurich, University Hospital Zurich), Prof. Andreas Papassotiropoulos (University of Basel), Prof. Ioannis Xenarios (Health2030 Genome Center), Dr. Bernd Rinn (ETH Zurich), Cornelia Kruschel Weber (University Hospital Zurich).
Additional persons involved: Dr. Marc Emmenegger, André Wethmar, Jacqueline Wiedler, Dr. Andra Chincisan, Leyla Batkitar, Lidia Madrigal, Laura Laks, Andrés Gonzalez Guerra, Andreia Delgado Magalhães, Dr. Chryssa Zografou, Dr. med. Katrin Frauenknecht, Jingjing Guo, Simone Hornemann, Marigona Imeri, Lisa Caflisch, Rea Müller, Antonella Rosati, Julie Domange, Dezirae Schneider , Magdalena Bialkowska, Lorene Mottier (University of Zurich, University Hospital Zurich); Prof. Dominique de Quervain, Dr. Thomas Schlitt, Dr. Pavlina Mastrandreas (University of Basel); Dr. David Lamparter, Dr Ilya Kolpakow (Health2030 Genome Center); Dr. Diana Coman Schmid, Simona Morello, Matti Veikko Johannes Heikkurinen, Cristian Scurtescu, Upal Nath, Michal Okoniewski, Dr. Franziskus Liem (ETH Zurich); Karin Edler, Patrick Hirschi, Katie Kalt (University Hospital Zurich).
Main achievements
Genomics and epigenetics can explain only a fraction of the human disease space. Mounting evidence suggests that many aspects of disease susceptibility are modulated by immune responses. The quantitative interrogation of human antibody repertoires – an important aspect of personalized health – and its correlation with medical data enables the discovery of novel pathogenic associations in humans.
We have used the infrastructures built during the SPHN funding period (see Figure 1) to conduct studies on ten thousands of hospital patients and healthy blood donors. For instance, we have studied the antibody profiles against the cellular prion protein and found that (1) natural anti-prion autoimmunity is likely innocuous and that (2) it might clear nascent prions early in life (Senatore et al., 2020). We also aimed to identify correlates of anti-tau autoimmunity and observed that antibodies against tau were not associated with neurodegeneration but with a novel nephro-vascular syndrome (Magalhães et al., 2021). High-throughput serology has been widely employed to respond to the SARS-CoV-2 pandemic. For example, we have investigated 72,105 individuals in the canton of Zurich from December 2019 to December 2020 and shown that the cumulative incidence rose to 12.5% by December 2020 (Emmenegger et al., 2020). In addition, we correlated medical and antibody data with the occurrence of antiphospholipid antibodies (Emmenegger, Saseendran Kumar, et al., 2021), and measured affinities in these samples (Denninger et al., 2021; Schneider et al., 2022). Our approach has resulted in unexpected findings that questioned previous research (Emmenegger, De Cecco, et al., 2021).
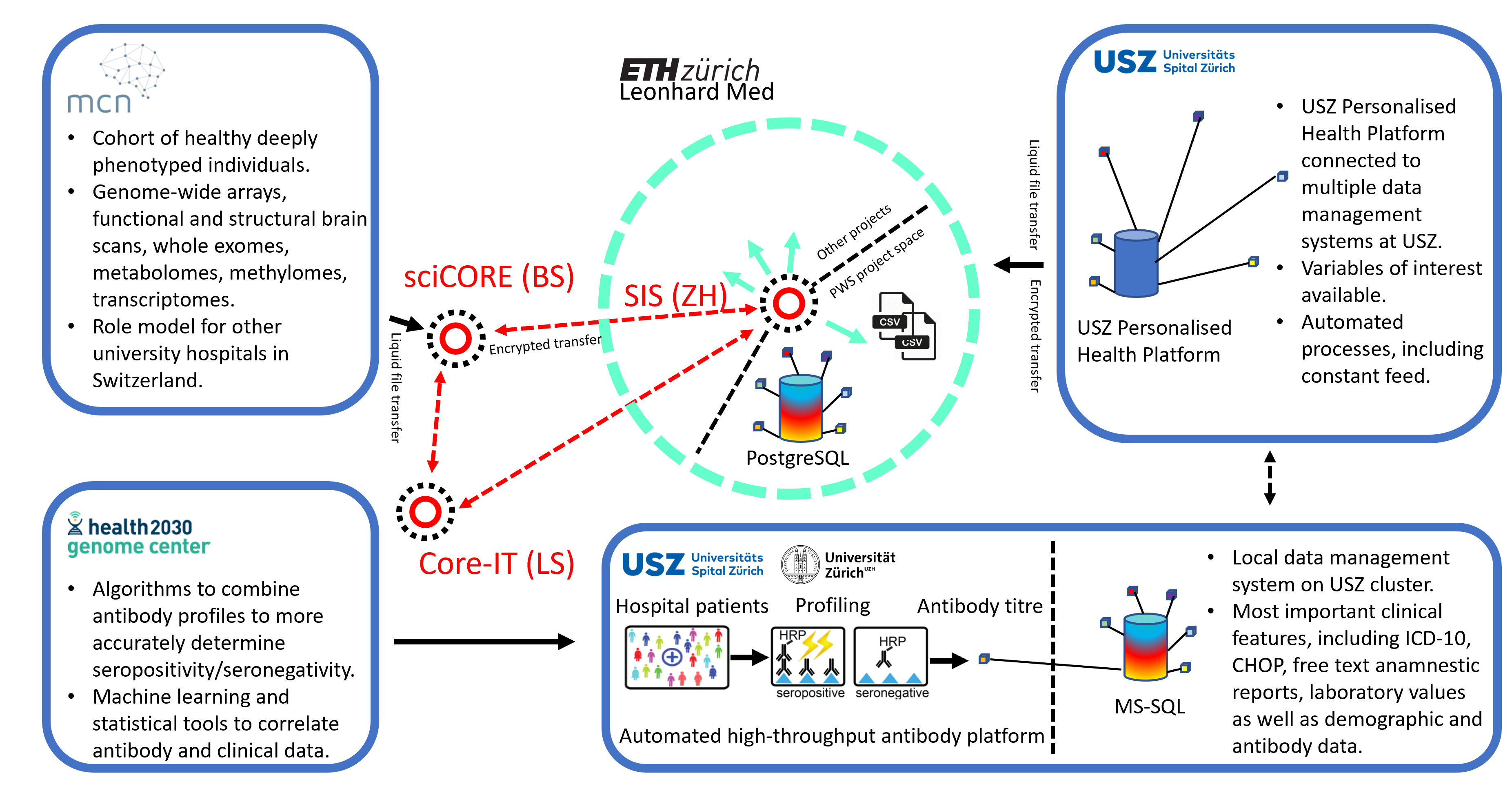
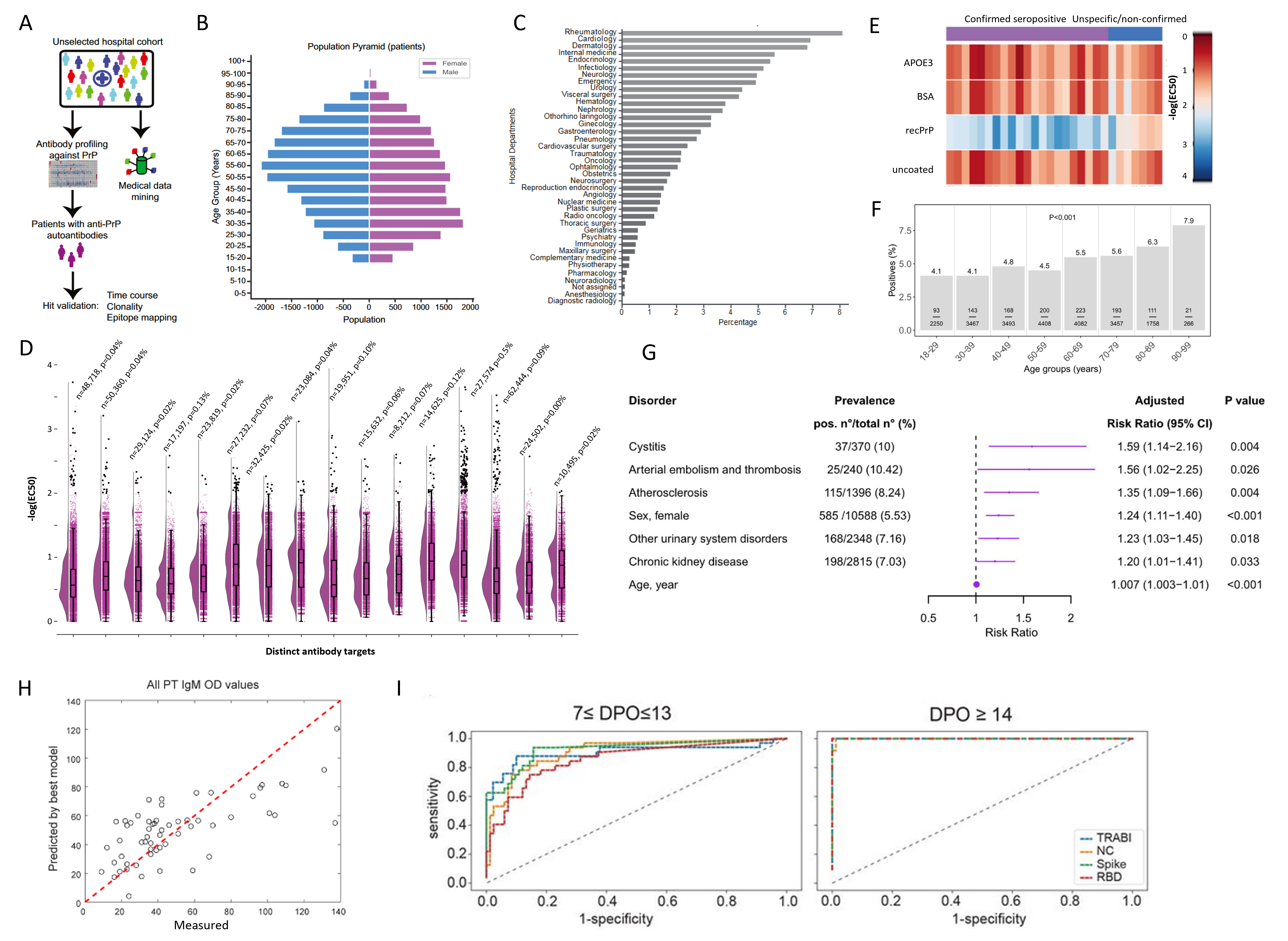
Figure 2. Summary of select results from antibody screens of the SPHN project. A. Workflow of the antibody profiling conducted on >30,000 patients against the cellular recombinant prion protein. B. From the same study as in (A), age and sex distribution. C. From the same study as in (A), the relative contribution of hospital departments to the samples included in the project. D. Antibody reactivity distributions (-log(EC50)) of 16 distinct targets. Black dots are considered seropositive samples where -log(EC50) exceeds a value of 2 and the error of the fit is less than 20% of the -log(EC50) value. n=number of samples screened. p=prevalence of seropositivity. E. Heatmap showing seropositive samples from same study as in (A) screened in a secondary assay for hit confirmation. F. Changes in antibody prevalence with age, seen for a screen performed with the microtubule-binding domain of the tau protein. G. A log-binominal regression model was used to infer clinical correlates of anti-tau seropositivity, resulting in the discovery of a nephro-vascular syndrome. H. The combined data of serological screening against SARS-CoV-2 proteins, anti-phospholipid antibodies, and clinical features led to the identification that anti-prothrombin IgM antibodies (PT IgM) emerge as a function of the strength of the antibody response against SARS-CoV2, with sex and disease severity as additional predictors in this linear mixed-effects model. I. The development and employment of a quadratic discriminatory analysis (QDA) enabled the combination of three independent measurements of SARS-CoV-2 proteins into one score, called TRABI. Shown are ROC curves. A-C, E: Data and graphs from (Senatore et al., 2020). F-G. Data and graphs from (Magalhães et al., 2021). H. Data and graphs from (Emmenegger, Saseendran Kumar, et al., 2021). I. Data and graphs from (Emmenegger et al., 2020).
Reusable infrastructure and datasets
Population-wide database with structured clinical and antibody data
We have created large datasets mainly, but not exclusively, consisting of pseudonymized health records of hospital patients from the University Hospital Zurich (> 200,000 samples), and the infrastructure to integrate, store, maintain, and analyse this data. Our main cohort represents a large subset of patients of an urban University Hospital center (with patients being included almost randomly based on surplus plasma material available for analysis in a project-specific high-throughput liquid biobank). This cohort is characterized by a plethora of disease conditions, the inclusion of almost all age groups (except children), and a diverse medical background. A part of the hospital registry collected for this SPHN project is mapped onto a pseudonym, which is then stored at the USZ in an MS-SQL database or, on Leonhard Med, in a Postgres database. Data from other sources – e.g. a cohort of oncology patients, patients with cardiometabolic diseases, or pediatric patients, or from other SPHN projects – can be joined on the project space and their samples can be investigated for the presence of antibodies against targets of choice. As a test case, our project has transferred data from the Transfaculty Research Platform Molecular and Cognitive Neurosciences (MCN), Basel. Apart from studying the relationship between the antibody repertoire and clinical/demographic data for which project-specific algorithms have been developed, the aggregation of such a wealth of data provides multiple opportunities: Research can be performed aiming at studying signatures other than the antibody repertoires and this data can be correlated with available clinical and demographic data. Or the clinical data itself can be used to investigate correlations among different disease entities.
Importantly, the cataloguing of data and a barrier-free access (using an easy-to-use web interface) as well as the sustainability of some of the project’s specific results can be improved by becoming a subproject of a large SPHN national data stream (NDS) project. Our consortium would welcome the integration of our infrastructure into a large ongoing SPHN initiative, which would be relatively simple having both DTUA and CA in place (for more information or to share our DTUA/CA, please contact us).
Available resources
Information on how the datasets and database can be accessed by contacting: Marc Emmenegger
Data dictionnaries
We work with CHOP, ATC, ICD, LOINC, where applicable. We use the data standards in use at the University Hospital Zurich. Triggered by this SPHN project, MCN has adapted these data standards for their data where applicable. A list of the specific data types can be shared with interested colleagues.
Watch the SPHN Webinar
Greater availability of sequencing technologies has accelerated the trend towards personalized medicine, most evident in clinical oncology. However, genome sequencing of cancer patients reflects only a small fraction of the potential of personalized medicine. In contrast, using our methodology, we are able to investigate the immune repertoire of large numbers of unselected individuals. While monoclonal antibodies obtained from humans may be utilized for the development of powerful and highly specific therapeutics, another important aspect is the ability to identify hidden disease associations by correlating the medical record with the individual antibody reactivity profile. We expect our pioneering approach to sustainably advance precision medicine and thus markedly promote personalized health in Switzerland.
References on Pubmed
- Pease D, Scheckel C, Schaper E, Eckhardt V, Emmenegger M, Xenarios I, Aguzzi A. Genome-wide identification of microRNAs regulating the human prion protein. Brain Pathol. 2019 Mar;29(2):232-244. doi: 10.1111/bpa.12679. Epub 2018 Dec 21. PMID: 30451334; PMCID: PMC8028613. https://doi.org/10.1111/bpa.12679
- Senatore A, Frontzek K, Emmenegger M, Chincisan A, Losa M, Reimann R, Horny G, Guo J, Fels S, Sorce S, Zhu C, George N, Ewert S, Pietzonka T, Hornemann S, Aguzzi A. Protective anti-prion antibodies in human immunoglobulin repertoires. EMBO Mol Med. 2020 Sep 7;12(9):e12739. doi: 10.15252/emmm.202012739. Epub 2020 Aug 10. PMID: 32776637; PMCID: PMC7506995.
- Emmenegger M, De Cecco E, Lamparater D et al. Early peak and rapid decline of SARS-CoV-2 seroprevalence in a Swiss metropolitan region. MedRxiv [Preprint], August 07, 2020. Available from: https://doi.org/10.1101/2020.05.31.20118554
- Cervia C, Nilsson J, Zurbuchen Y, Valaperti A, Schreiner J, Wolfensberger A, Raeber ME, Adamo S, Weigang S, Emmenegger M, Hasler S, Bosshard PP, De Cecco E, Bächli E, Rudiger A, Stüssi-Helbling M, Huber LC, Zinkernagel AS, Schaer DJ, Aguzzi A, Kochs G, Held U, Probst-Müller E, Rampini SK, Boyman O. Systemic and mucosal antibody responses specific to SARS-CoV-2 during mild versus severe COVID-19. J Allergy Clin Immunol. 2021 Feb;147(2):545-557.e9. doi: 10.1016/j.jaci.2020.10.040. Epub 2020 Nov 20. PMID: 33221383; PMCID: PMC7677074.
- Avar M, Heinzer D, Steinke N, Doğançay B, Moos R, Lugan S, Cosenza C, Hornemann S, Andréoletti O, Aguzzi A. Prion infection, transmission, and cytopathology modeled in a low-biohazard human cell line. Life Sci Alliance. 2020 Jun 30;3(8):e202000814. doi: 10.26508/lsa.202000814. PMID: 32606072; PMCID: PMC7335386.
- Emmenegger M, De Cecco E, Hruska-Plochan M, Eninger T, Schneider MM, Barth M, Tantardini E, de Rossi P, Bacioglu M, Langston RG, Kaganovich A, Bengoa-Vergniory N, Gonzalez-Guerra A, Avar M, Heinzer D, Reimann R, Häsler LM, Herling TW, Matharu NS, Landeck N, Luk K, Melki R, Kahle PJ, Hornemann S, Knowles TPJ, Cookson MR, Polymenidou M, Jucker M, Aguzzi A. LAG3 is not expressed in human and murine neurons and does not modulate α-synucleinopathies. EMBO Mol Med. 2021 Sep 7;13(9):e14745. doi: 10.15252/emmm.202114745. Epub 2021 Jul 26. PMID: 34309222; PMCID: PMC8422075.
- Heinzer D, Avar M, Pease DP, Dhingra A, Yin JA, Schaper E, Doğançay B, Emmenegger M, Spinelli A, Maggi K, Chincisan A, Mead S, Hornemann S, Heutink P, Aguzzi A. Novel regulators of PrPC biosynthesis revealed by genome-wide RNA interference. PLoS Pathog. 2021 Oct 27;17(10):e1010013. doi: 10.1371/journal.ppat.1010013. PMID: 34705895; PMCID: PMC8575309.
- Schneider MM, Emmenegger M, Xu CK, Condado Morales I, Meisl G, Turelli P, Zografou C, Zimmermann MR, Frey BM, Fiedler S, Denninger V, Jacquat RP, Madrigal L, Ilsley A, Kosmoliaptsis V, Fiegler H, Trono D, Knowles TP, Aguzzi A. Microfluidic characterisation reveals broad range of SARS-CoV-2 antibody affinity in human plasma. Life Sci Alliance. 2021 Nov 30;5(2):e202101270. doi: 10.26508/lsa.202101270. PMID: 34848436; PMCID: PMC8645332.
- Emmenegger M, Kumar SS, Emmenegger V, Malinauskas T, Buettner T, Rose L, Schierack P, Sprinzl MF, Sommer CJ, Lackner KJ, Aguzzi A, Roggenbuck D, Frauenknecht KBM. Anti-prothrombin autoantibodies enriched after infection with SARS-CoV-2 and influenced by strength of antibody response against SARS-CoV-2 proteins. PLoS Pathog. 2021 Dec 3;17(12):e1010118. doi: 10.1371/journal.ppat.1010118. PMID: 34860860; PMCID: PMC8673606.
- Magalhães AD, Emmenegger M, De Cecco E, Carta M, Frontzek K, Chincisan A, Hornemann S, Aguzzi A. Large-scale seroepidemiology identifies a nephro-vascular syndrome associated with autoimmune reactivity to tau.
medRxiv [Preprint], November 26, 2021. Available from: https://doi.org/10.1101/2021.11.24.21266833 - Müller M, Avar M, Heinzer D, Emmenegger M, Aguzzi A, Pelkmans L, Berry S. High content genome-wide siRNA screen to investigate the coordination of cell size and RNA production. Sci Data. 2021 Jun 28;8(1):162. doi: 10.1038/s41597-021-00944-5. Erratum in: Sci Data. 2021 Dec 2;8(1):313. PMID: 34183683; PMCID: PMC8239010.
- Fiedler S, Denninger V, Morgunov AS, Ilsley A, Worth R, Meisl G, Xu CK, Piziorska MA, Ricci R, Malik AY, Devenish SRA, Schneider MM, Kosmoliaptsis V, Aguzzi A, Iwasaki A, Fiegler H, Knowles TPJ. Mutations in two SARS-CoV-2 variants of concern reflect two distinct strategies of antibody escape. bioRxiv [Preprint], July 26, 2021. Available from: https://doi.org/10.1101/2021.07.23.453327
- Kirschenbaum D, Voigt FF, Dadgar-Kiani E, Catto F, Trevisan C, Bichsel O, Shirani H, Nilsson PR, Frontzek KJ, Paganetti P, Helmchen F, Lee JH, Aguzzi A. Local predictors of anti-Aβ therapy efficacy revealed by quantitative whole-brain microscopy. bioRxiv [Preprint], July 14, 2021. Available from: https://doi.org/10.1101/2021.01.15.426090
- Carta M, Aguzzi A. Molecular foundations of prion strain diversity. Curr Opin Neurobiol. 2021 Jul 27;72:22-31. doi: 10.1016/j.conb.2021.07.010. Epub ahead of print. PMID: 34416480.
- Fiedler S, Devenish SRA, Morgunov AS, Ilsley A, Ricci F, Emmenegger M, Kosmoliaptsis V, Theel ES, Mills JR, Sholukh AM, Aguzzi A, Iwasaki A, Lynn AK, Knowles TPJ. Serological fingerprints link antiviral activity of therapeutic antibodies to affinity and concentration. bioRxiv [Preprint], February 9, 2022. Available from: https://doi.org/10.1101/2022.02.03.478946.
-
Gehlhausen JR, Little AJ, Ko CJ, Emmenegger M, Lucas C, Wong P, Klein J, Lu P, Mao T, Jaycox J, Wang E; Yale IMPACT Team, Ugwu N, Muenker C, Mekael D, Klein RQ, Patrignelli R, Antaya R, McNiff J, Damsky W, Kamath K, Shon J, Ring AM, Yildirim I, Omer S, Ko AI, Aguzzi A, Iwasaki A. Lack of association between pandemic chilblains and SARS-CoV-2 infection. Proc Natl Acad Sci U S A. 2022 Mar 1;119(9):e2122090119. doi: 10.1073/pnas.2122090119. PMID: 35217624; PMCID: PMC8892496.
- Denninger V, Xu CK, Meisl G, Morgunov AS, Fiedler S, Ilsley A, Emmenegger M, Malik AY, Piziorska MA, Schneider MM, Devenish SRA, Kosmoliaptsis V, Aguzzi A, Fiegler H, Knowles TPJ. Microfluidic Antibody Affinity Profiling Reveals the Role of Memory Reactivation and Cross-Reactivity in the Defense Against SARS-CoV-2. ACS Infect Dis. 2022 Mar 30. doi: 10.1021/acsinfecdis.1c00486. Epub ahead of print. PMID: 35352558.
Disclaimer: The contents on this website are intended as a general source of information and have been provided by the project PIs. The SPHN Management Office is not responsible for its accuracy, validity, or completeness.

