Swiss PharmacoKinetics clinical data warehouse (SwissPKcdw)
Project consortium: Prof. Stefanie Krämer (ETH Zürich), Prof. Christoph Berger (University Children's Hospital Zurich), Dr. Paolo Paioni (University Children's Hospital Zurich), Prof. Henriette Meyer zu Schwabendissen (University of Basel), Dr. Bernd Rinn (ETH Zürich), Dr. Diana Coman Schmid (ETH Zürich), Dr. Oliver Renn (ETH Zürich).
Additional persons involved: Beat Bangerter (Division of Infectious Diseases and Hospital Epidemiology, University Children's Hospital Zürich), Diana Coman-Schmid (Scientific IT Services, ETH Zürich), Dr. Roland Goers (Biopharmacy, University Basel), Vera F. Jäggi (Division of Infectious Diseases and Hospital Epidemiology, University Children's Hospital Zürich), Dr. Daniel Müller (Institute of Clinical Chemistry, University Hospital Zurich), Dr. Althea Parker (Scientific IT Services, ETH Zürich), Dr. Thierry Sengstag, Center for Scientific Computing, University of Basel), Romy Tilen (Division of Infectious Diseases and Hospital Epidemiology, University Children's Hospital Zürich), Swen Vermeul (Scientific IT Services, ETH Zürich, Zürich), Dr. med. Julia Bielicki (Paediatric Infectious Diseases, University of Basel Children’s Hospital).
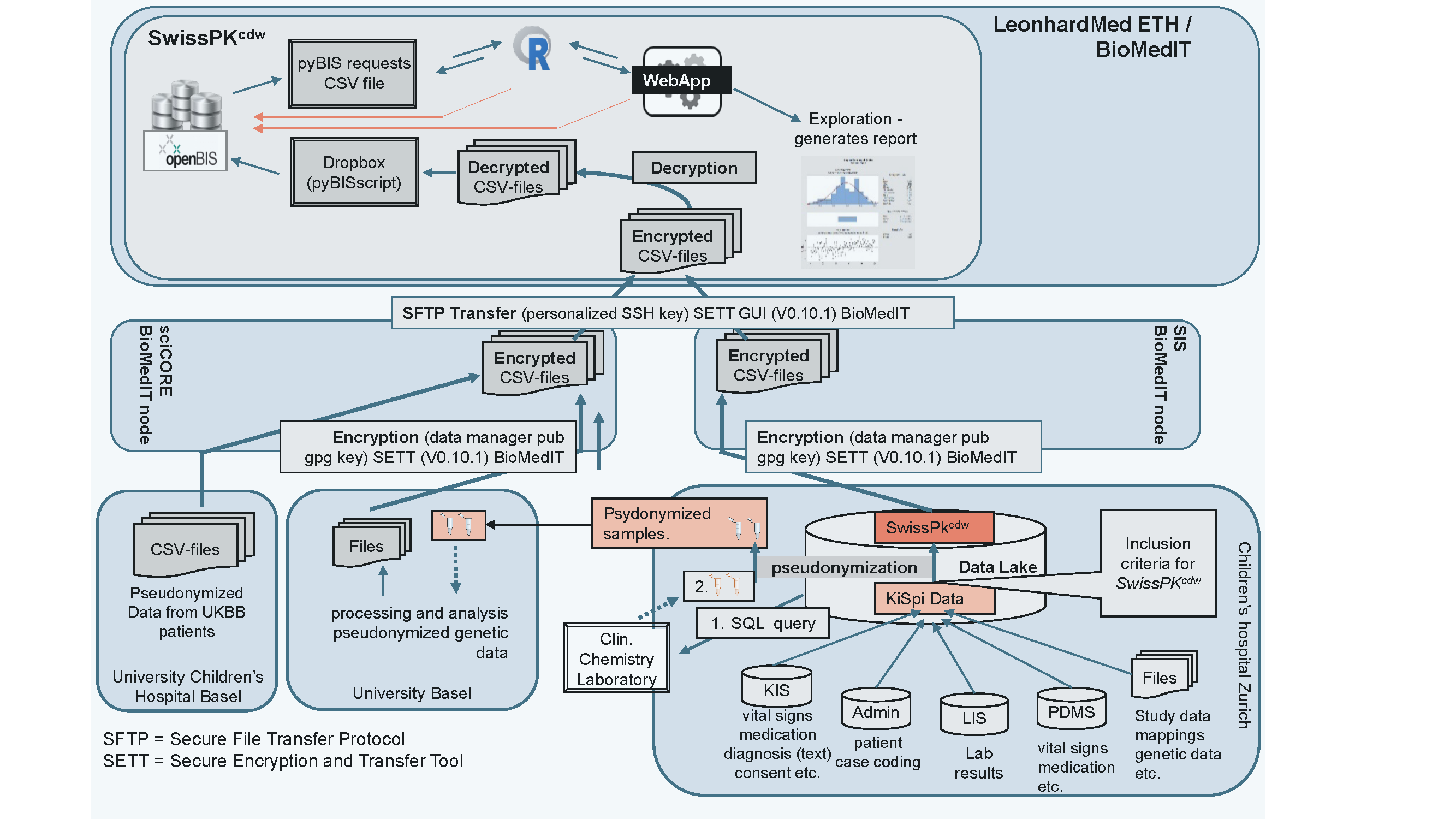
Figure 1. The SwissPKcdw infrastructure
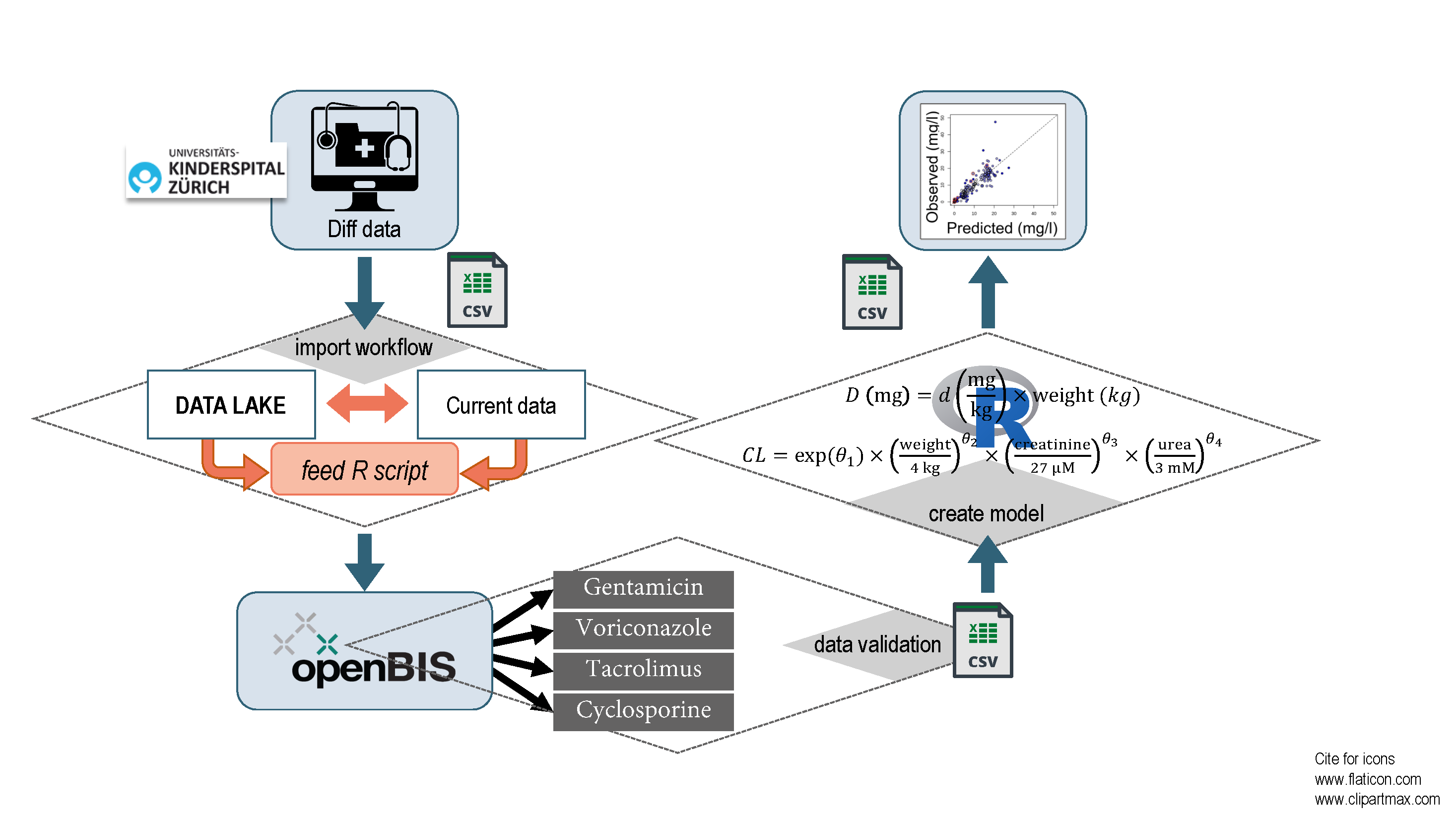
Figure 2. Summary of Workflows for the import, management and analysis of the data.
Main achievements
The main achievement of this infrastructure development project is the establishment of the clinical data warehouse (CDW) and data analysis platform Swiss PharmacoKinetics clinical data warehouse (SwissPKcdw).
The SwissPKcdw provides the means for safe transfer, storage and management of pseudonymized patient data from two children's hospitals (Zurich and Basel) and the University of Basel (research laboratory data). The SwissPKcdw platform is designed to follow the FAIR principles. It contains open source software for data analysis and provides access to the data for population pharmacokinetic modelling and optimization of dosing schemes. The CDW and modelling platform have successfully been implemented in the secure environment of Leonhard Med at the BioMedIT node Zurich. The Data Transfer and Use Agreement (DTUA) including Consortium Agreement (CA), the Data Transfer and Processing Agreement (DTPA), the Material Transfer Agreement (MTA) and the user policy are in place. Clinical data from the hospitals with corresponding pharmacogenetic data generated in the Biopharmacy within the Department of Pharmaceutical Sciences, University of Basel, were successfully transferred to the SwissPKcdw tenant, stored, managed and analyzed by population pharmacokinetics modelling. The data in the CDW can be explored with a dedicated web application within the secure environment.
Reusable infrastructure and datasets
Clinical data warehouse (SwissPKcdw) on LeonhardMed
Figure 1 provides an overview of the SwissPKcdw infrastructure, including the dataflow from the source hospitals to the SwissPKcdw tenant on Leonhard Med at the BioMedIT node Zurich. The basic IT infrastructures (at hospitals and BioMedIT), data governance (Data Transfer and Use Agreement (DTUA), Data Transfer and Processing Agreement (DTPA) and protocols for the data flow, management and analysis were established during the SwissPKcdw project (Figure 1). All data are transferred, stored and processed within the BioMedIT, in compliance with the SPHN Information Security Policy.
Data mapping, harmonization and pseudonymization are performed at the source before encryption and transfer using the Secure Encryption and Transfer Tool (SETT). Data transfer is coordinated by the Data Coordination Center (DCC) of BioMedIT. Access to SwissPKcdw is restricted to authorized users. Security controls are imposed at infrastructure and policy level. The data are harmonized following the recommendations of the SPHN semantic interoperability framework (SPHN dataset).
The SwissPKcdw platform has a modular structure for data management (DM), analysis (DA) and exploration (DE). Figure 2 summarizes the workflows for the import, management and analysis of the data with the final export of anonymized results. The DM is built around openBIS, a versatile system for data management and secure handling of confidential data. For data import into openBIS, a customized tool reads the raw SQL database dumps, standardizes and validates the data and reports possibly invalid data. The new data is compared via an intermediate SQLite database with already imported data, creating new or updating existing entries. Finally, the data are imported and assigned to the correct space. In openBIS, each individual patient and the related data are associated with a unique research identifier or pseudonym. All entries and their changes are traceable by versioning.
The DA module includes software for data analysis. To connect the DM with the DA module, a Python script was developed, which queries for all data in a given openBIS space and exports the data to a file for analysis. Finally, the SwissPKcdw web application provides graphical user access to predefined statistical analyses (module DA) that are applied to data managed with openBIS (module DM) and offers exploration functionalities (module DE) for selected parameters (text adapted from Goers, Coman Schmid et al., 2021).
Available resources
- Contact: Dr. Paolo Paioni, University Children’s Hospital Zurich, Switzerland
- Table 1. Collected data and corresponding data structure (Download)
- DTUA (Download)
- MTA (Download)
Modeling platform within SwissPKcdw
The SwissPKcdw tenant on LeonhardMed contains a space for data analysis. The open source software R (with R Studio), Python and Jupyter Notebook are available for data analysis. The user with granted access can install R packages in the user space. Parallel computing is available with up to 36 processors, enabling complex data analysis by population pharmacokinetics modelling. The results of the analysis, devoid of any personal or identifiable data, can be transferred from the safe environment to the local environment via Gitlab or copy paste with a respective user interface(e.g., MobaXterm).
Available resources
- Contact: Dr. Paolo Paioni, University Children’s Hospital Zurich, Switzerland
- Users access the modeling platform via the secure login to LeonhardMed with two factor authentication.
- SwissPKcdw User Policy (Download)
- Gitlab for exemplary R scripts for population pharmacokinetics modeling with the R package saemix (Link)
SwissPKcdw web application
The SwissPKcdw web application provides graphical user access to predefined statistical analyses (from the data analysis module) that are applied to data managed with openBIS (data management module) and currently offers exploration functionalities (data exploration module) for selected parameters (e.g., patient cohort, medication, clinical variables). The web application is implemented in JavaScript and uses openCPU
Available resources
- Users access the web app via the secure login to LeonhardMed remote desktop with two factor authentication.
- Contact: Dr. Paolo Paioni, University Children’s Hospital Zurich, Switzerland
Watch the SPHN Webinar
To enhance access to clinical data from a high number of paediatric patients seen at the hospitals, the project will establish a clinical data warehouse (CDW). It will collect drug plasma concentrations from routine drug monitoring and clinical trials of paediatric patients together with clinical and prescription data as well as genetic information. CDW data are transferred to be analysed and shared on a connected platform. This data is essential to establish mathematic modelling, to eventually mirror the physiological processes that affect the drug in the developing and growing body of a child (pharmacokinetics). With this so-called pharmacokinetic modelling, dosage regimens for children can be optimised.
References on Pubmed
- Goers, Coman Schmid et al., SwissPKcdw - A clinical data warehouse for the optimization of pediatric dosing regimens. CPT Pharmacometrics Syst. Pharmacol. 2021 (in press). doi: 10.1002/psp4.12723.
- Paioni et al., Gentamicin population pharmacokinetics in pediatric patients-A prospective study with data analysis using the saemix package in R. Pharmaceutics. 2021;13:1596. doi: 10.3390/pharmaceutics13101596
Disclaimer: The contents on this website are intended as a general source of information and have been provided by the project PIs. The SPHN Management Office is not responsible for its accuracy, validity, or completeness.

