SPO: Swiss Personalized Oncology
Project consortium: Main PIs: Prof. Olivier Michielin (CHUV) and Prof. Mohamed Bentires-Alj (UniBS)
Pierre-Yves Dietrich (HUG), Adrian Ochsenbein (Inselspital), Georges Coukos (CHUV), Christian Lovis (UniGE), Oliver Verscheure (SDSC, EPFL, ETHZ), Mark Rubin (UniBE), Jacques Fellay (CHUV), Patrick Ruch (HEG/HES-SO), Michel Cuendet (CHUV), Sylvain Pradervand (CHUV), Petros Tsantoulis (HUG), Krisztian Homicsko (CHUV), Roger von Moos (SAKK, Kantonsspital Graubünden), Markus Manz (USZ), Andreas Wicki (USB), Mitch Levesque (USZ), Christian Britschgi (USZ), Walter Weber (USB), Luigi Terracciano (UniBS/USB)
Main achievements
The SPO Driver project co-led by Prof. Olivier Michielin and Prof. Mohamed Bentires-Alj established a consortium to make clinical and molecular data interoperability within Switzerland, addressing patients from in- and out-patient clinics. The SPO project federates clinical and -omics data acquisition, standardization, and use for precision oncology within Switzerland. As such, it involves not only the five university hospitals, but also non-University structures under the umbrella of the Swiss Group for Clinical Cancer Research (SAKK).
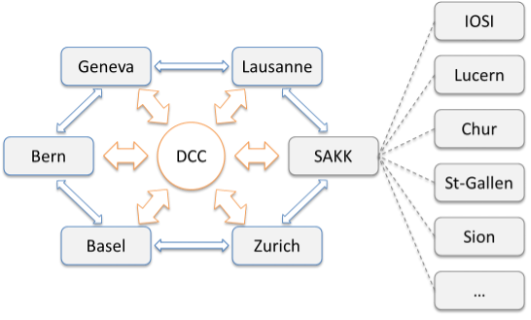
Figure 1: Structure of the Swiss Network for the Personalized Oncology Driver Project. All university centres are interconnected via the Data Coordination Center (DCC). SAKK acts as an additional hub, aggregating the data from regional centers.
We defined a core clinical data set to capture the essential phenotype for personalized oncology. This dataset was defined and modeled in compliance with the guidelines of the SPHN semantic interoperability framework.
In order to test the newly established data infrastructures of SPHN, we set up a pilot project in the context of a research question ‘Outcome analysis of Swiss cancer patients exhibiting a BRAF non-V600 mutation in their tumor or liquid biopsies, in comparison to patients with a BRAF p.V600 mutation’. This pilot study has the potential to make a significant contribution to the clinical management of patients with BRAF non-V600 mutated malignancies.
Based on SPO core dataset, the SAKK has been building up sustainable processes and infrastructures for collecting real world data in oncology from diagnosis to loss of follow-up: The Swiss Centralized Oncology Real-world Evidence Data (SCORED) initiative. A joint SPHN/Oncology (SPO) – SAKK steering committee, composed of representatives of both SAKK and the SPO Consortium, has been set up to discuss and evaluate research project applications requesting access to SCORED data.
A monthly national molecular tumor board has been established. Patient cases are presented and possible strategies concerning further diagnostic procedures or the next line of therapy are discussed. In addition, this board has proven to be useful to exchange information on running trials in individual centers, thus paving the way to refer patients for specific molecularly matched trials to a site with a suitable active study.
SPO has also established a set of Standard Operating Procedures to harmonize live-cell biobanking across Switzerland. These protocols will enable the interoperability of Swiss oncology biobanks and ensure larger, high-quality cohorts for further investigation and discovery-based applications.
Reusable infrastructures and datasets
SPO Core Data Set
The SPO dataset was properly defined and modeled in compliance with the guidelines of the SPHN semantic interoperability framework. Links to standards like SNOMED-CT were implemented when possible. We ensured that it was fully aligned with international initiatives like ICGC-ARGO and mCODE.
These specifications were used to build a cohort of patients with a mutation of the BRAF gene in their tumor (SPO pilot project) and a cohort of melanoma patients. They provide also a blue-print for the SAKK SCORED registry core data set definition.
The SPO core dataset includes the following categories:
- Demographics
- Oncological diagnosis
- Staging / Grading
- Laboratory
- Somatic variants
- Treatments (drug, radiotherapy, surgery)
- Outcome
- Follow-up / Survival status
Available resources
Variables are now incorporated in the SPHN reference dataset (https://git.dcc.sib.swiss/sphn-semantic-framework/sphn-ontology) and in the SPO specific ontology (https://git.dcc.sib.swiss/hospital-it/project-sphn-spo).
Contact: Sylvain Pradervand, Lausanne University Hospital (CHUV), sylvain.pradervand@chuv.ch
An automated pipeline to standardize RDF data reception on Biomed-IT with quality control, SPARQL loading and reporting
The RDF format is designed to ensure compatibility from different data providers. However, these data are often, if not always, mapped from original relational data tables. As these tables follow internal logic at each university hospital, one expects to find some semantic variations or missing attributes resulting from uncompleted mapping. To ensure semantic harmonization, the description of concepts and concept relations, referred to as ontology or metadata, are defined in a central repository. Alongside harmonized ontology, a set of semantic rules written in shapes constraint language (SHACL) are available in the same repository. An erroneous mapping of the original data can generate practical errors that are highly context-specific and that can pass the SHACL validation. For instance, this kind of error can consist of patients missing oncology-related diagnoses or treatments. Therefore, to ensure interoperability, we designed a data validation pipeline that will handle data at their reception on BioMedIT with the least possible number of manual operations. The pipeline will validate the data against the correct ontology version, report the errors and load a RDF database.
Available resources
The pipeline code is available at: https://git.dcc.sib.swiss/spo/data-reception.
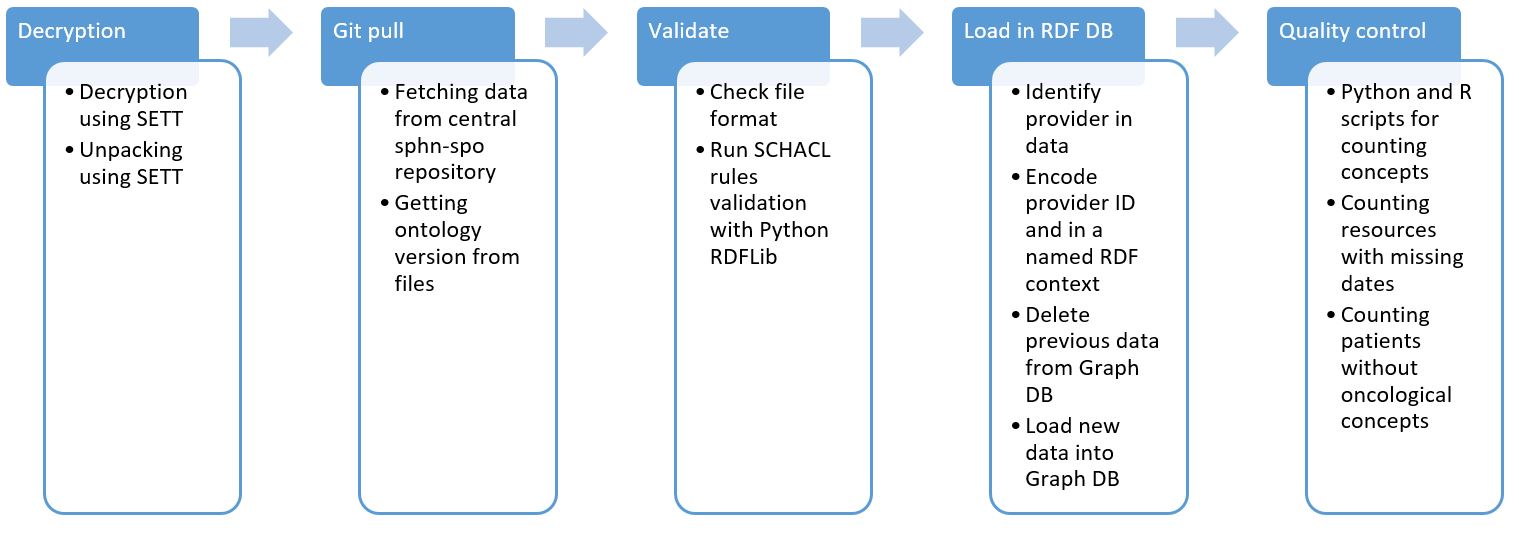
Figure 2: Overview of data processing and validation pipeline
A Natural Language Processing tool that extracts a treatment response label given a radiology report conclusion
Response Evaluation Criteria in Solid Tumors (RECIST) is the main instrument to monitor the treatment response of solid tumors. These criteria are mainly determined by radiological investigations and they are legacy to evaluate the clinical outcomes of clinical trials. However, RECIST criteria are rarely used for routine oncological treatments and therefore RECIST are not commonly found in Electronic Health Records. The objective of the SPO text mining working group was the exploration of advanced Artificial Intelligence and Natural Language Processing techniques to support the automatic assignment of the four RECIST scales (Disease Progression, Stable Disease, Partial Response, Complete Response) based on radiology reports. We also aim at evaluating how languages and institutional specificities of five large teaching hospitals are likely to affect the quality of the RECIST categorization in French and German language. In our approach, 7 machine learning methods were evaluated to establish a strong baseline. The best strategies yield average F1-scores of 90% and 85.8% respectively for the 2-classes classification (Progressive/Non-progressive disease) and the 4-classes classification (RECIST). These results are approaching the manual assignment of RECIST labels as measured by Matthew’s correlation coefficient and Cohen’s Kappa (79% and 76%). On this basis, we confirm the capacity of specific models to generalize on new unseen data and we assess the impact of using pre-trained language models and transformers on the accuracy of the classifiers. Hence, the performances of automatic RECIST labelling tackle human classification despite the residual ambiguity within the wording of radiology reports.
Available resources
The software is deployed on BioMedIT within SPO project space. The software will be deployed in University Hospital upon request.
The Swiss Centralized Oncology Real-world Evidence Data (SCORED) database
The SAKK SCORED initiative focuses on establishing an infrastructure for registry projects to fight cancer by providing nationwide evidence of clinical practice in strong collaboration with all partners of the health care system. The captured data will be available to multiple partners, first and foremost to the researchers of the participating centers of the SAKK network (22 cancer treatment centers).
SCORED projects are designed to create highest benefit and development in data science in oncology by collecting interoperable data that can be shared between projects. The SCORED infrastructure comprises data from SCORED registry projects, which will then also be available for future retrospective projects. SCORED registry projects must collect at least the SCORED/SPO core data set. If required, registry projects can also collect additional variables. The core data set and the additionally collected variables of specific registry projects is stored at SAKK in the SCORED data warehouse. The data is analyzed for the specific registry project and is available for future retrospective projects tackling a new research question.
Available resources
SCORED database information can be requested through the SAKK SCORED team.
Contact person is the Project Manager: Daniel.Hugelshofer@sakk.ch
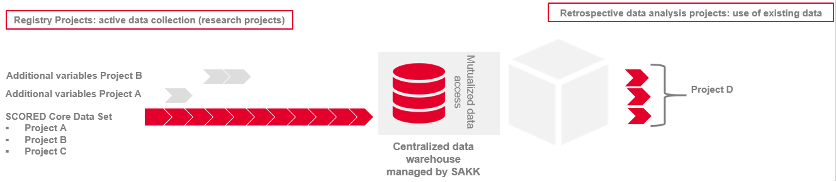
Figure 3: Data interoperability for SCORED. If patients are eligible for several registries, their data will be captured once and will be used for all approved projects (red blocks).
The Swiss Molecular Tumor Board (SMTB)
The SMTB has been established in the first phase of SPO between the five university hospitals and takes place regularly via remote conference. Difficult cases are discussed, with all centers sharing their experience and giving crucial feedback on treatment opportunities, including information on active clinical trials.
In the second phase of SPO (SPO NDS), the national molecular tumor board is now going to be coupled to a PHRT-supported biological testing program. Thus, it will evolve into a platform for diagnostic testing and therapy proposals beyond standard of care. Results from the testing program will flow back into the national molecular tumor board and will be used to (i) inform therapy decisions where appropriate and (ii) to feed a detailed and granular database with deep information on tumor biology, applied therapies and outcome. This database will be mined for diagnostic, prognostic and predictive information with the aim of developing machine-learning based algorithms for therapy prediction.
Available resources
The SMTB is run by all participating centres. Central coordination is provided by the clinical workgroup of SPO NDS. Contacts:
- Andreas Wicki, University and University Hospital Zurich (andreas.wicki@usz.ch)
- Laura Boos, University and University Hospital Zurich (boos@usz.ch)
- Cristina Golfieri, University of Basel (cristina.golfieri@unibas.ch)
Standardized protocols for sample handling that creates standards for tissue processing across Switzerland
Together with the SAKK we developed a protocol manual for sample handling that creates standards for tissue processing across Switzerland. The manual includes those we developed within SPO (i.e., live-cell tumor freezing and live-cell peripheral blood mononuclear cell preservation), as well as specific procedures for biobanks to generate up to 30 other materials for research. This effort will ensure that all oncology clinics in Switzerland have access to the same standard operating procedures that will help facilitate sharing of materials and pooling of cohorts regardless of which hospital recruits the patient.
To facilitate the analysis of retrospective cancer cohorts, we have built several tissue microarrays (TMAs) that enable the simultaneous immunohistochemical analysis of dozens to hundreds of patients. TMAs of up to 900 melanoma patients were developed for this project.
Available resources
- Protocol manual is available upon request from the SAKK and will be shared across all university hospitals and participating hospitals across Switzerland.
- Publication established best-practices and scientific validation of protocols for international scientific audience.
- Tissue Microarrays are stable and usable for collaboration for any academic researcher with appropriate ethical permission.
Data dictionnaries
| Data type | Concept name | Ontology |
| Clinical routine | Administrative Gender | SPHN |
| Clinical routine | Adverse Event | SPHN |
| Clinical routine | Birth Date | SPHN |
| Clinical routine | Body Height | SPHN |
| Clinical routine | Body Site | SPHN |
| Clinical routine | Body Weight | SPHN |
| Clinical routine | Code | SPHN |
| Clinical routine | Death Status | SPHN |
| Clinical routine | Drug | SPHN |
| Clinical routine | Drug Administration Event | SPHN |
| Clinical routine | Duration | SPHN |
| Clinical routine | Follow Up Event | SPO |
| Clinical routine | FOPH Diagnosis | SPHN |
| Clinical routine | FOPH Procedure | SPHN |
| Clinical routine | Frequency | SPHN |
| Clinical routine | Oncology Drug Treatment | SPHN |
| Clinical routine | Oncology Surgery | SPHN |
| Clinical routine | Oncology Treatment Assessment | SPHN |
| Clinical routine | Oncology Treatment Assessment | SPO |
| Clinical routine | Radiology Report | SPHN |
| Clinical routine | Radiotherapy Procedure | SPHN |
| Clinical routine | Regimen Component | SPO |
| Clinical routine | Relative Dose of Drug Applied | SPHN |
| Clinical routine | Simple Score | SPHN |
| Clinical routine | Substance | SPHN |
| Clinical routine | Substance Amount | SPHN |
| Clinical routine | Time Pattern | SPHN |
| Clinical routine | Treatment Cycle | SPO |
| Clinical routine | Unit | SPHN |
| Diagnostics | Biobanksample | SPHN |
| Diagnostics | Biosample | SPHN |
| Diagnostics | ICD-O Diagnosis | SPHN |
| Diagnostics | Laterality | SPO |
| Diagnostics | Oncology Diagnosis | SPO |
| Diagnostics | TNM Classification | SPHN |
| Diagnostics | Tumor Grade | SPHN |
| Diagnostics | Tumor Specimen | SPO |
| Diagnostics | Tumor Stage | SPHN |
| Diagnostics / Genetic | Genetic Region Studied | SPO |
| Diagnostics / Genetic | Genetic Test | SPO |
| Diagnostics / Genetic | Genetic Variant Found | SPO |
| General consent | Consent | SPHN |
| Laboratory | Lab Result | SPHN |
Follow up projects – continuation – next steps
The existing infrastructures, datasets, and know-how developed over the 3 years of SPO project will be further extended through an SPHN National Data Stream funding (SPO-NDS). The SPO-NDS will consolidate the clinical data acquisition at the 5 university hospitals and at the Swiss Group for Clinical Cancer Research (SAKK). High value cohorts will be assembled in cancer types treated with immuno-oncology (IO) therapies that will allow comparative immunomics between highly IO sensitive tumors (e.g., melanoma, lung-cancer, triple-negative breast cancer) to less sensitive (e.g., MSI-CRC, hormone receptor-positive breast cancer). Using the momentum of the Tumor Profiler (TuPro) initiative, single-cell and -omics data will be at the center of the program including NGS, CyTOF/IMC, digital pathology, as well as functional drug screening to drive therapeutic decisions within the existing molecular tumor boards. The patients will readily benefit from these efforts as the infrastructure and the -omics data will be directly employed to support treatment decisions within the molecular tumor boards nationwide.
Watch the SPHN webinar
References on Pubmed
- Raisaro JL, Troncoso-Pastoriza JR, Pradervand S, Cuendet M, Misbach M, Sa J, Marino F, Freundler N, Rosat N, Cavin D, Leichtle A, Fellay J, Michielin O, Hubaux JP. SPHN/PHRT - MedCo in Action: Empowering the Swiss Molecular Tumor Board with Privacy-Preserving and Real-Time Patient Discovery. Stud Health Technol Inform. 2020 Jun 16;270:1161-1162. doi: 10.3233/SHTI200345. PMID: 32570563.
-
Ghisoni E, Wicky A, Bouchaab H, Imbimbo M, Delyon J, Gautron Moura B, Gérard CL, Latifyan S, Özdemir BC, Caikovski M, Pradervand S, Tavazzi E, Gatta R, Marandino L, Valabrega G, Aglietta M, Obeid M, Homicsko K, Mederos Alfonso NN, Zimmermann S, Coukos G, Peters S, Cuendet MA, Di Maio M, Michielin O. Late-onset and long-lasting immune-related adverse events from immune checkpoint-inhibitors: An overlooked aspect in immunotherapy. Eur J Cancer. 2021 May;149:153-164. doi: 10.1016/j.ejca.2021.03.010. Epub 2021 Apr 14. PMID: 33865201.
Disclaimer: The contents on this website are intended as a general source of information and have been provided by the project PIs. The SPHN Management Office is not responsible for its accuracy, validity, or completeness.

